Role of chemokine receptor CXCR2 expression in mammary tumor growth, angiogenesis and metastasis
- PMID: 22368515
- PMCID: PMC3284109
- DOI: 10.4103/1477-3163.92308
Role of chemokine receptor CXCR2 expression in mammary tumor growth, angiogenesis and metastasis
Abstract
Background: Chemokines and their receptors have long been known to regulate metastasis in various cancers. Previous studies have shown that CXCR2 expression is upregulated in malignant breast cancer tissues but not in benign ductal epithelial samples. The functional role of CXCR2 in the metastatic phenotype of breast cancer still remains unclear. We hypothesize that the chemokine receptor, CXCR2, mediates tumor cell invasion and migration and promotes metastasis in breast cancer. The objective of this study is to investigate the potential role of CXCR2 in the metastatic phenotype of mouse mammary tumor cells.
Materials and methods: We evaluated the functional role of CXCR2 in breast cancer by stably downregulating the expression of CXCR2 in metastatic mammary tumor cell lines Cl66 and 4T1, using short hairpin RNA (shRNA). The effects of CXCR2 downregulation on tumor growth, invasion and metastatic potential were analyzed in vitro and in vivo.
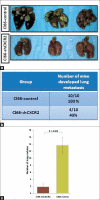
Results: We demonstrated knock down of CXCR2 in Cl66 and 4T1 cells (Cl66-shCXCR2 and 4T1-shCXCR2) cells by reverse transcriptase polymerase chain reaction (RT-PCR) at the transcriptional level and by immunohistochemistry at the protein level. We did not observe a significant difference in in vitro cell proliferation between vector control and CXCR2 knock-down Cl66 or 4T1 cells. Next, we examined the invasive potential of Cl66-shCXCR2 cells by in vitro Matrigel invasion assay. We observed a significantly lower number (52 ± 5) of Cl66-shCXCR2 cells invading through Matrigel compared to control cells (Cl66-control) (182 ± 3) (P < 0.05). We analyzed the in vivo metastatic potential of Cl66-shCXCR2 using a spontaneous metastasis model by orthotopically implanting cells into the mammary fat pad of female BALB/c mice. Animals were sacrificed 12 weeks post tumor implantation and tissue samples were analyzed for metastatic nodules. CXCR2 downregulation significantly inhibited tumor cell metastasis. All the mice (n = 10) implanted with control Cl66 cells spontaneously developed lung metastasis, whereas a significantly lower number of mice (40%) implanted with Cl66-shCXCR2 cells exhibited lung metastases.
Conclusions: Together, these results suggest that CXCR2 may play a critical role in breast cancer invasion and metastasis.
Keywords: CXC chemokines; CXCR2; metastasis; tumor growth.
Figures




Similar articles
-
CXCR2: A Novel Mediator of Mammary Tumor Bone Metastasis.Int J Mol Sci. 2019 Mar 12;20(5):1237. doi: 10.3390/ijms20051237. Int J Mol Sci. 2019. PMID: 30871004 Free PMC article.
-
IL-17-CXC Chemokine Receptor 2 Axis Facilitates Breast Cancer Progression by Up-Regulating Neutrophil Recruitment.Am J Pathol. 2020 Jan;190(1):222-233. doi: 10.1016/j.ajpath.2019.09.016. Epub 2019 Oct 22. Am J Pathol. 2020. PMID: 31654638 Free PMC article.
-
Host Cxcr2-dependent regulation of mammary tumor growth and metastasis.Clin Exp Metastasis. 2015 Jan;32(1):65-72. doi: 10.1007/s10585-014-9691-0. Epub 2014 Dec 16. Clin Exp Metastasis. 2015. PMID: 25511644 Free PMC article.
-
Small interfering RNA-mediated CXCR1 or CXCR2 knock-down inhibits melanoma tumor growth and invasion.Int J Cancer. 2010 Jan 15;126(2):328-36. doi: 10.1002/ijc.24714. Int J Cancer. 2010. PMID: 19585580 Free PMC article.
-
Targeting CXCR2 enhances chemotherapeutic response, inhibits mammary tumor growth, angiogenesis, and lung metastasis.Mol Cancer Ther. 2013 May;12(5):799-808. doi: 10.1158/1535-7163.MCT-12-0529. Epub 2013 Mar 6. Mol Cancer Ther. 2013. PMID: 23468530 Free PMC article.
Cited by
-
High CXCR2 expression predicts poor prognosis in adult patients with acute myeloid leukemia.Ther Adv Hematol. 2020 Sep 14;11:2040620720958586. doi: 10.1177/2040620720958586. eCollection 2020. Ther Adv Hematol. 2020. PMID: 32973988 Free PMC article.
-
Targeting Angiogenesis in Breast Cancer: Current Evidence and Future Perspectives of Novel Anti-Angiogenic Approaches.Front Pharmacol. 2022 Feb 25;13:838133. doi: 10.3389/fphar.2022.838133. eCollection 2022. Front Pharmacol. 2022. PMID: 35281942 Free PMC article. Review.
-
L1CAM deployed perivascular tumor niche promotes vessel wall invasion of tumor thrombus and metastasis of renal cell carcinoma.Cell Death Discov. 2023 Apr 4;9(1):112. doi: 10.1038/s41420-023-01410-4. Cell Death Discov. 2023. PMID: 37015905 Free PMC article.
-
Conditioning solid tumor microenvironment through inflammatory chemokines and S100 family proteins.Cancer Lett. 2015 Aug 28;365(1):11-22. doi: 10.1016/j.canlet.2015.05.002. Epub 2015 May 8. Cancer Lett. 2015. PMID: 25963887 Free PMC article. Review.
-
High throughput screening of cytokines, chemokines and matrix metalloproteinases in wound fluid induced by mammary surgery.Oncotarget. 2015 Oct 6;6(30):29296-310. doi: 10.18632/oncotarget.4828. Oncotarget. 2015. PMID: 26313265 Free PMC article.
References
-
- Murphy PM, Tiffany HL. Cloning of complementary DNA encoding a functional human interleukin-8 receptor. Science. 1991;253:1280–3. - PubMed
-
- Ahuja SK, Murphy PM. The CXC chemokines growth-regulated oncogene (GRO) alpha, GRObeta, GROgamma, neutrophil-activating peptide-2, and epithelial cell-derived neutrophil-activating peptide-78 are potent agonists for the type B, but not the type A, human interleukin-8 receptor. J Biol Chem. 1996;271:20545–50. - PubMed
-
- Ahuja SK, Lee JC, Murphy PM. CXC chemokines bind to unique sets of selectivity determinants that can function independently and are broadly distributed on multiple domains of human interleukin-8 receptor B.Determinants of high affinity binding and receptor activation are distinct. J Biol Chem. 1996;271:225–32. - PubMed
-
- Fan X, Patera AC, Pong-Kennedy A, Deno G, Gonsiorek W, Manfra DJ, et al. Murine CXCR1 is a functional receptor for GCP-2/CXCL6 and interleukin-8/CXCL8. J Biol Chem. 2007;282:11658–66. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

