Analysis of the transcriptome of Panax notoginseng root uncovers putative triterpene saponin-biosynthetic genes and genetic markers
- PMID: 22369100
- PMCID: PMC3287501
- DOI: 10.1186/1471-2164-12-S5-S5
Analysis of the transcriptome of Panax notoginseng root uncovers putative triterpene saponin-biosynthetic genes and genetic markers
Abstract
Background: Panax notoginseng (Burk) F.H. Chen is important medicinal plant of the Araliacease family. Triterpene saponins are the bioactive constituents in P. notoginseng. However, available genomic information regarding this plant is limited. Moreover, details of triterpene saponin biosynthesis in the Panax species are largely unknown.
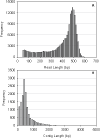
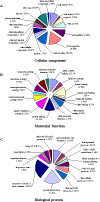
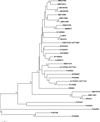
Results: Using the 454 pyrosequencing technology, a one-quarter GS FLX titanium run resulted in 188,185 reads with an average length of 410 bases for P. notoginseng root. These reads were processed and assembled by 454 GS De Novo Assembler software into 30,852 unique sequences. A total of 70.2% of unique sequences were annotated by Basic Local Alignment Search Tool (BLAST) similarity searches against public sequence databases. The Kyoto Encyclopedia of Genes and Genomes (KEGG) assignment discovered 41 unique sequences representing 11 genes involved in triterpene saponin backbone biosynthesis in the 454-EST dataset. In particular, the transcript encoding dammarenediol synthase (DS), which is the first committed enzyme in the biosynthetic pathway of major triterpene saponins, is highly expressed in the root of four-year-old P. notoginseng. It is worth emphasizing that the candidate cytochrome P450 (Pn02132 and Pn00158) and UDP-glycosyltransferase (Pn00082) gene most likely to be involved in hydroxylation or glycosylation of aglycones for triterpene saponin biosynthesis were discovered from 174 cytochrome P450s and 242 glycosyltransferases by phylogenetic analysis, respectively. Putative transcription factors were detected in 906 unique sequences, including Myb, homeobox, WRKY, basic helix-loop-helix (bHLH), and other family proteins. Additionally, a total of 2,772 simple sequence repeat (SSR) were identified from 2,361 unique sequences, of which, di-nucleotide motifs were the most abundant motif.
Conclusion: This study is the first to present a large-scale EST dataset for P. notoginseng root acquired by next-generation sequencing (NGS) technology. The candidate genes involved in triterpene saponin biosynthesis, including the putative CYP450s and UGTs, were obtained in this study. Additionally, the identification of SSRs provided plenty of genetic makers for molecular breeding and genetics applications in this species. These data will provide information on gene discovery, transcriptional regulation and marker-assisted selection for P. notoginseng. The dataset establishes an important foundation for the study with the purpose of ensuring adequate drug resources for this species.
Figures



References
-
- Wang CZ, McEntee E, Wicks S, Wu JA, Yuan CS. Phytochemical and analytical studies of Panax notoginseng (Burk.) F.H.Chen. J Nat Med. 2006;60:97–106. doi: 10.1007/s11418-005-0027-x. - DOI
-
- Ng TB. Pharmacological activity of sanchi ginseng (Panax notoginseng) J Pharm Pharmacol. 2006;58(8):1007–1019. - PubMed
-
- Wu W, Zhang XM, Liu PM, Li JM, Wang JF. Effects of Panax notoginseng saponin Rg1 on cardiac electrophysiological properties and ventricular fibrillation threshold in dogs. Acta Pharmacol Sin. 1995;16(5):459–463. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

