Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay
- PMID: 22371084
- PMCID: PMC3297981
- DOI: 10.1038/nbt.2137
Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay
Abstract
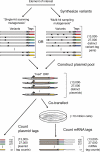
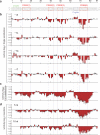
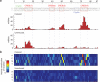
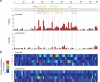
Learning to read and write the transcriptional regulatory code is of central importance to progress in genetic analysis and engineering. Here we describe a massively parallel reporter assay (MPRA) that facilitates the systematic dissection of transcriptional regulatory elements. In MPRA, microarray-synthesized DNA regulatory elements and unique sequence tags are cloned into plasmids to generate a library of reporter constructs. These constructs are transfected into cells and tag expression is assayed by high-throughput sequencing. We apply MPRA to compare >27,000 variants of two inducible enhancers in human cells: a synthetic cAMP-regulated enhancer and the virus-inducible interferon-β enhancer. We first show that the resulting data define accurate maps of functional transcription factor binding sites in both enhancers at single-nucleotide resolution. We then use the data to train quantitative sequence-activity models (QSAMs) of the two enhancers. We show that QSAMs from two cellular states can be combined to design enhancer variants that optimize potentially conflicting objectives, such as maximizing induced activity while minimizing basal activity.
Figures

Comment in
-
In-depth functional dissection of enhancers.Nat Methods. 2012 Apr;9(4):322-3. doi: 10.1038/nmeth.1963. Nat Methods. 2012. PMID: 22563600 No abstract available.
-
Dissecting genomic regulatory elements in vivo.Nat Biotechnol. 2012 Jun 7;30(6):504-6. doi: 10.1038/nbt.2266. Nat Biotechnol. 2012. PMID: 22678387 No abstract available.
References
-
- Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. - PubMed
-
- Dorer DE, Nettelbeck DM. Targeting cancer by transcriptional control in cancer gene therapy and viral oncolysis. Adv Drug Deliv Rev. 2009;61:554–571. - PubMed
-
- Fan F, Wood KV. Bioluminescent assays for high-throughput screening. Assay Drug Dev Technol. 2007;5:127–136. doi:10.1089/adt.2006.053. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

