Rapid genetic change underpins antagonistic coevolution in a natural host-pathogen metapopulation
- PMID: 22372578
- PMCID: PMC3319837
- DOI: 10.1111/j.1461-0248.2012.01749.x
Rapid genetic change underpins antagonistic coevolution in a natural host-pathogen metapopulation
Abstract
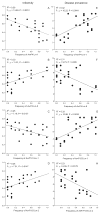
Antagonistic coevolution is a critical force driving the evolution of diversity, yet the selective processes underpinning reciprocal adaptive changes in nature are not well understood. Local adaptation studies demonstrate partner impacts on fitness and adaptive change, but do not directly expose genetic processes predicted by theory. Specifically, we have little knowledge of the relative importance of fluctuating selection vs. arms-race dynamics in maintaining polymorphism in natural systems where metapopulation processes predominate. We conducted cross-year epidemiological, infection and genetic studies of multiple wild host and pathogen populations in the Linum-Melampsora association. We observed asynchronous phenotypic fluctuations in resistance and infectivity among demes. Importantly, changes in allelic frequencies at pathogen infectivity loci, and in host recognition of these genetic variants, correlated with disease prevalence during natural epidemics. These data strongly support reciprocal coevolution maintaining balanced resistance and infectivity polymorphisms, and highlight the importance of characterising spatial and temporal dynamics in antagonistic interactions.
© 2012 Blackwell Publishing Ltd/CNRS.
Figures
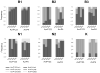
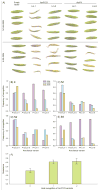
 =2004–2006;
=2004–2006;
 =2006–2008) and infectivity (
=2006–2008) and infectivity (
 =2002–2004;
=2002–2004;
 =2004–2006;
=2004–2006;
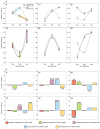
 =2006–2008). (B) Changes in infectivity and resistance between consecutive time periods (see text for details). Significant changes are highlighted (*<0.05, **<0.01, ***<0.001, ****<0.0001; see Table S1).
=2006–2008). (B) Changes in infectivity and resistance between consecutive time periods (see text for details). Significant changes are highlighted (*<0.05, **<0.01, ***<0.001, ****<0.0001; see Table S1).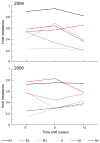
References
-
- Altermatt F, Ebert D. Genetic diversity of Daphnia magna populations enhances resistance to parasites. Ecol Lett. 2008;11:918–928. - PubMed
-
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26:32–46.
-
- Bergelson J, Kreitman M, Stahl EA, Tian D. Evolutionary dynamics of plant R-genes. Science. 2001;292:2281–2285. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

