Development, evaluation and application of 3D QSAR Pharmacophore model in the discovery of potential human renin inhibitors
- PMID: 22372967
- PMCID: PMC3287469
- DOI: 10.1186/1471-2105-12-S14-S4
Development, evaluation and application of 3D QSAR Pharmacophore model in the discovery of potential human renin inhibitors
Abstract
Background: Renin has become an attractive target in controlling hypertension because of the high specificity towards its only substrate, angiotensinogen. The conversion of angiotensinogen to angiotensin I is the first and rate-limiting step of renin-angiotensin system and thus designing inhibitors to block this step is focused in this study.
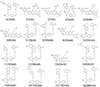
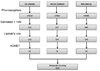
Methods: Ligand-based quantitative pharmacophore modeling methodology was used in identifying the important molecular chemical features present in the set of already known active compounds and the missing features from the set of inactive compounds. A training set containing 18 compounds including active and inactive compounds with a substantial degree of diversity was used in developing the pharmacophore models. A test set containing 93 compounds, Fischer randomization, and leave-one-out methods were used in the validation of the pharmacophore model. Database screening was performed using the best pharmacophore model as a 3D structural query. Molecular docking and density functional theory calculations were used to select the hit compounds with strong molecular interactions and favorable electronic features.
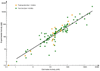
Results: The best quantitative pharmacophore model selected was made of one hydrophobic, one hydrogen bond donor, and two hydrogen bond acceptor features with high a correlation value of 0.944. Upon validation using an external test set of 93 compounds, Fischer randomization, and leave-one-out methods, this model was used in database screening to identify chemical compounds containing the identified pharmacophoric features. Molecular docking and density functional theory studies have confirmed that the identified hits possess the essential binding characteristics and electronic properties of potent inhibitors.
Conclusion: A quantitative pharmacophore model of predictive ability was developed with essential molecular features of a potent renin inhibitor. Using this pharmacophore model, two potential inhibitory leads were identified to be used in designing novel and future renin inhibitors as antihypertensive drugs.
Figures
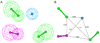


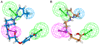
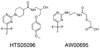

Similar articles
-
Prediction of Novel Anoctamin1 (ANO1) Inhibitors Using 3D-QSAR Pharmacophore Modeling and Molecular Docking.Int J Mol Sci. 2018 Oct 17;19(10):3204. doi: 10.3390/ijms19103204. Int J Mol Sci. 2018. PMID: 30336555 Free PMC article.
-
Theoretical investigation on known renin inhibitors and generation of ligand-based pharmacophore models for hypertension treatment.J Biomol Struct Dyn. 2024;42(24):13411-13420. doi: 10.1080/07391102.2023.2275186. Epub 2023 Oct 28. J Biomol Struct Dyn. 2024. PMID: 37897186
-
Exploration of the structural requirements of HIV-protease inhibitors using pharmacophore, virtual screening and molecular docking approaches for lead identification.J Mol Graph Model. 2015 Mar;56:20-30. doi: 10.1016/j.jmgm.2014.11.015. Epub 2014 Dec 5. J Mol Graph Model. 2015. PMID: 25541527
-
Potent BACE-1 inhibitor design using pharmacophore modeling, in silico screening and molecular docking studies.BMC Bioinformatics. 2011 Feb 15;12 Suppl 1(Suppl 1):S28. doi: 10.1186/1471-2105-12-S1-S28. BMC Bioinformatics. 2011. PMID: 21342558 Free PMC article.
-
Identification of Potent Small-Molecule PCSK9 Inhibitors Based on Quantitative Structure-Activity Relationship, Pharmacophore Modeling, and Molecular Docking Procedure.Curr Probl Cardiol. 2023 Jun;48(6):101660. doi: 10.1016/j.cpcardiol.2023.101660. Epub 2023 Feb 24. Curr Probl Cardiol. 2023. PMID: 36841313 Review.
Cited by
-
Topology-enhanced molecular graph representation for anti-breast cancer drug selection.BMC Bioinformatics. 2022 Sep 19;23(1):382. doi: 10.1186/s12859-022-04913-6. BMC Bioinformatics. 2022. PMID: 36123643 Free PMC article.
-
Pharmacophore modeling and virtual screening for the discovery of new type 4 cAMP phosphodiesterase (PDE4) inhibitors.PLoS One. 2013 Dec 10;8(12):e82360. doi: 10.1371/journal.pone.0082360. eCollection 2013. PLoS One. 2013. PMID: 24340020 Free PMC article.
-
Integration of Ligand-Based and Structure-Based Methods for the Design of Small-Molecule TLR7 Antagonists.Molecules. 2022 Jun 23;27(13):4026. doi: 10.3390/molecules27134026. Molecules. 2022. PMID: 35807273 Free PMC article.
-
3D-QSAR-Based Pharmacophore Modeling, Virtual Screening, and Molecular Dynamics Simulations for the Identification of Spleen Tyrosine Kinase Inhibitors.Front Cell Infect Microbiol. 2022 Jun 30;12:909111. doi: 10.3389/fcimb.2022.909111. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35846777 Free PMC article.
-
Tubulin inhibitors: pharmacophore modeling, virtual screening and molecular docking.Acta Pharmacol Sin. 2014 Jul;35(7):967-79. doi: 10.1038/aps.2014.34. Epub 2014 Jun 9. Acta Pharmacol Sin. 2014. PMID: 24909516 Free PMC article.
References
-
- Yasuchika Y, Keith M, Nissim CC, Robert M, Frédéric C, Christian S, Jeanette MW, Jürgen M. The P1 N-isopropyl motif bearing hydroxyethylene dipeptide isostere analogues of aliskiren are in vitro potent inhibitors of the human aspartyl protease renin. Bio org Med Chem Lett. 2009;19:4863–4867. doi: 10.1016/j.bmcl.2009.05.128. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

