Transparent mediation-based access to multiple yeast data sources using an ontology driven interface
- PMID: 22372975
- PMCID: PMC3471333
- DOI: 10.1186/1471-2105-13-S1-S7
Transparent mediation-based access to multiple yeast data sources using an ontology driven interface
Abstract
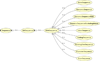
Background: Saccharomyces cerevisiae is recognized as a model system representing a simple eukaryote whose genome can be easily manipulated. Information solicited by scientists on its biological entities (Proteins, Genes, RNAs...) is scattered within several data sources like SGD, Yeastract, CYGD-MIPS, BioGrid, PhosphoGrid, etc. Because of the heterogeneity of these sources, querying them separately and then manually combining the returned results is a complex and time-consuming task for biologists most of whom are not bioinformatics expert. It also reduces and limits the use that can be made on the available data.
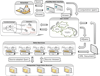
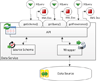
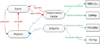
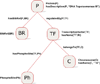
Results: To provide transparent and simultaneous access to yeast sources, we have developed YeastMed: an XML and mediator-based system. In this paper, we present our approach in developing this system which takes advantage of SB-KOM to perform the query transformation needed and a set of Data Services to reach the integrated data sources. The system is composed of a set of modules that depend heavily on XML and Semantic Web technologies. User queries are expressed in terms of a domain ontology through a simple form-based web interface.
Conclusions: YeastMed is the first mediation-based system specific for integrating yeast data sources. It was conceived mainly to help biologists to find simultaneously relevant data from multiple data sources. It has a biologist-friendly interface easy to use. The system is available at http://www.khaos.uma.es/yeastmed/.
Figures

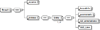



Similar articles
-
Federated ontology-based queries over cancer data.BMC Bioinformatics. 2012 Jan 25;13 Suppl 1(Suppl 1):S9. doi: 10.1186/1471-2105-13-S1-S9. BMC Bioinformatics. 2012. PMID: 22373043 Free PMC article.
-
KA-SB: from data integration to large scale reasoning.BMC Bioinformatics. 2009 Oct 1;10 Suppl 10(Suppl 10):S5. doi: 10.1186/1471-2105-10-S10-S5. BMC Bioinformatics. 2009. PMID: 19796402 Free PMC article.
-
YEASTRACT: providing a programmatic access to curated transcriptional regulatory associations in Saccharomyces cerevisiae through a web services interface.Nucleic Acids Res. 2011 Jan;39(Database issue):D136-40. doi: 10.1093/nar/gkq964. Epub 2010 Oct 23. Nucleic Acids Res. 2011. PMID: 20972212 Free PMC article.
-
Evolution of web services in bioinformatics.Brief Bioinform. 2005 Jun;6(2):178-88. doi: 10.1093/bib/6.2.178. Brief Bioinform. 2005. PMID: 15975226 Review.
-
The ontology of craniofacial development and malformation for translational craniofacial research.Am J Med Genet C Semin Med Genet. 2013 Nov;163C(4):232-45. doi: 10.1002/ajmg.c.31377. Epub 2013 Oct 4. Am J Med Genet C Semin Med Genet. 2013. PMID: 24124010 Free PMC article. Review.
Cited by
-
The PhosphoGRID Saccharomyces cerevisiae protein phosphorylation site database: version 2.0 update.Database (Oxford). 2013 May 13;2013:bat026. doi: 10.1093/database/bat026. Print 2013. Database (Oxford). 2013. PMID: 23674503 Free PMC article.
References
-
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. Life with 6000 genes. Science. 1996;274(546):563–547. - PubMed
-
- Lambrix P, Jakoniene V. In: Proceedings of the First Asia-Pacific bioinformatics conference on Bioinformatics; 4-7 February 2003; Adelaide, Australia. Chen Y-PP, editor. Australian Computer Society, Inc; 2003. Towards transparent access to multiple biological databanks; pp. 53–60.
-
- Hernandez T, Kambhampati S. Integration of biological sources: current systems and challenges ahead. SIGMOD Rec. 2004;33:51–60.
-
- Davidson SB, Crabtree J, Brunk BP, Schug J, Tannen V, Overton GC, Stoeckert JCJ. K2/Kleisli and GUS: experiments in integrated access to Genomic Data Sources. IBM System Journal. 2001;40:512–531.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

