Federated ontology-based queries over cancer data
- PMID: 22373043
- PMCID: PMC3471355
- DOI: 10.1186/1471-2105-13-S1-S9
Federated ontology-based queries over cancer data
Abstract
Background: Personalised medicine provides patients with treatments that are specific to their genetic profiles. It requires efficient data sharing of disparate data types across a variety of scientific disciplines, such as molecular biology, pathology, radiology and clinical practice. Personalised medicine aims to offer the safest and most effective therapeutic strategy based on the gene variations of each subject. In particular, this is valid in oncology, where knowledge about genetic mutations has already led to new therapies. Current molecular biology techniques (microarrays, proteomics, epigenetic technology and improved DNA sequencing technology) enable better characterisation of cancer tumours. The vast amounts of data, however, coupled with the use of different terms - or semantic heterogeneity - in each discipline makes the retrieval and integration of information difficult.
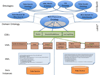
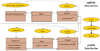
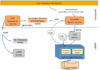
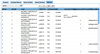
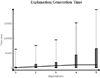
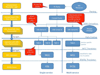
Results: Existing software infrastructures for data-sharing in the cancer domain, such as caGrid, support access to distributed information. caGrid follows a service-oriented model-driven architecture. Each data source in caGrid is associated with metadata at increasing levels of abstraction, including syntactic, structural, reference and domain metadata. The domain metadata consists of ontology-based annotations associated with the structural information of each data source. However, caGrid's current querying functionality is given at the structural metadata level, without capitalising on the ontology-based annotations. This paper presents the design of and theoretical foundations for distributed ontology-based queries over cancer research data. Concept-based queries are reformulated to the target query language, where join conditions between multiple data sources are found by exploiting the semantic annotations. The system has been implemented, as a proof of concept, over the caGrid infrastructure. The approach is applicable to other model-driven architectures. A graphical user interface has been developed, supporting ontology-based queries over caGrid data sources. An extensive evaluation of the query reformulation technique is included.
Conclusions: To support personalised medicine in oncology, it is crucial to retrieve and integrate molecular, pathology, radiology and clinical data in an efficient manner. The semantic heterogeneity of the data makes this a challenging task. Ontologies provide a formal framework to support querying and integration. This paper provides an ontology-based solution for querying distributed databases over service-oriented, model-driven infrastructures.
Figures





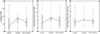

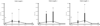
Similar articles
-
A semantic proteomics dashboard (SemPoD) for data management in translational research.BMC Syst Biol. 2012;6 Suppl 3(Suppl 3):S20. doi: 10.1186/1752-0509-6-S3-S20. Epub 2012 Dec 17. BMC Syst Biol. 2012. PMID: 23282161 Free PMC article.
-
BioFed: federated query processing over life sciences linked open data.J Biomed Semantics. 2017 Mar 15;8(1):13. doi: 10.1186/s13326-017-0118-0. J Biomed Semantics. 2017. PMID: 28298238 Free PMC article.
-
Gauging triple stores with actual biological data.BMC Bioinformatics. 2012 Jan 25;13 Suppl 1(Suppl 1):S3. doi: 10.1186/1471-2105-13-S1-S3. BMC Bioinformatics. 2012. PMID: 22373359 Free PMC article.
-
Towards Semantic e-Science for Traditional Chinese Medicine.BMC Bioinformatics. 2007 May 9;8 Suppl 3(Suppl 3):S6. doi: 10.1186/1471-2105-8-S3-S6. BMC Bioinformatics. 2007. PMID: 17493289 Free PMC article. Review.
-
Techniques for optimization of queries on integrated biological resources.J Bioinform Comput Biol. 2004 Jun;2(2):375-411. doi: 10.1142/s0219720004000648. J Bioinform Comput Biol. 2004. PMID: 15297988 Review.
Cited by
-
A unified structural/terminological interoperability framework based on LexEVS: application to TRANSFoRm.J Am Med Inform Assoc. 2013 Sep-Oct;20(5):986-94. doi: 10.1136/amiajnl-2012-001312. Epub 2013 Apr 9. J Am Med Inform Assoc. 2013. PMID: 23571850 Free PMC article.
-
Ontologies and Knowledge Graphs in Oncology Research.Cancers (Basel). 2022 Apr 10;14(8):1906. doi: 10.3390/cancers14081906. Cancers (Basel). 2022. PMID: 35454813 Free PMC article. Review.
-
Electronic Health Record-Oriented Knowledge Graph System for Collaborative Clinical Decision Support Using Multicenter Fragmented Medical Data: Design and Application Study.J Med Internet Res. 2024 Jul 5;26:e54263. doi: 10.2196/54263. J Med Internet Res. 2024. PMID: 38968598 Free PMC article.
-
Defragged Binary I Ching Genetic Code Chromosomes Compared to Nirenberg's and Transformed into Rotating 2D Circles and Squares and into a 3D 100% Symmetrical Tetrahedron Coupled to a Functional One to Discern Start From Non-Start Methionines through a Stella Octangula.J Proteome Sci Comput Biol. 2012;2012(1):3. doi: 10.7243/2050-2273-1-3. J Proteome Sci Comput Biol. 2012. PMID: 23431415 Free PMC article.
-
Cancer bioinformatics: a new approach to systems clinical medicine.BMC Bioinformatics. 2012 May 1;13:71. doi: 10.1186/1471-2105-13-71. BMC Bioinformatics. 2012. PMID: 22549015 Free PMC article. No abstract available.
References
-
- NCRI Informatics Initiative. http://www.cancerinformatics.org.uk/
-
- caBIG® Programme. https://cabig.nci.nih.gov/
-
- Tobias J, Chilukuri R, Komatsoulis GA, Mohanty S, Sioutos N, Warzel DB, Wright LW, Crowley RS. The CAP cancer protocols-a case study of caCORE based data standards implementation to integrate with the Cancer Biomedical Informatics Grid. BMC Med Inform Decis Mak. 2006;6:25–25. doi: 10.1186/1472-6947-6-25. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

