Identification of ubiquitin/ubiquitin-like protein modification from tandem mass spectra with various PTMs
- PMID: 22373085
- PMCID: PMC3287473
- DOI: 10.1186/1471-2105-12-S14-S8
Identification of ubiquitin/ubiquitin-like protein modification from tandem mass spectra with various PTMs
Abstract
Background: Various solutions have been introduced for the identification of post-translational modification (PTM) from tandem mass spectrometry (MS/MS) in proteomics field but the identification of peptide modifiers, such as Ubiquitin (Ub) and ubiquitin-like proteins (Ubls), is still a challenge. The fragmentation of peptide modifier produce complex shifted ion mass patterns in combination with other PTMs, which makes it difficult to identify and locate the PTMs on a protein sequence. Currently, most PTM identification methods do not consider the complex fragmentation of peptide modifier or deals it separately from the other PTMs.
Results: We developed an advanced PTM identification method that inspects possible ion patterns of the most known peptide modifiers as well as other known biological and chemical PTMs to make more comprehensive and accurate conclusion. The proposed method searches all detectable mass differences of measured peaks from their theoretical values and the mass differences within mass tolerance range are grouped as mass shift classes. The most possible locations of multiple PTMs including peptide modifiers can be determined by evaluating all possible scenarios generated by the combination of the qualified mass shift classes.The proposed method showed excellent performance in the test with simulated spectra having various PTMs including peptide modifiers and in the comparison with recently developed methods such as QuickMod and SUMmOn. In the analysis of HUPO Brain Proteome Project (BPP) datasets, the proposed method could find the ubiquitin modification sites that were not identified by other conventional methods.
Conclusions: This work presents a novel method for identifying bothpeptide modifiers that generate complex fragmentation patternsand PTMs that are not fragmented during fragmentation processfrom tandem mass spectra.
Figures
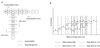



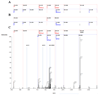
Similar articles
-
Automated identification of SUMOylation sites using mass spectrometry and SUMmOn pattern recognition software.Nat Methods. 2006 Jul;3(7):533-9. doi: 10.1038/nmeth891. Nat Methods. 2006. PMID: 16791211
-
An accurate and efficient algorithm for Peptide and ptm identification by tandem mass spectrometry.Genome Inform. 2007;19:119-30. Genome Inform. 2007. PMID: 18546510
-
The utility of ETD mass spectrometry in proteomic analysis.Biochim Biophys Acta. 2006 Dec;1764(12):1811-22. doi: 10.1016/j.bbapap.2006.10.003. Epub 2006 Oct 30. Biochim Biophys Acta. 2006. PMID: 17118725 Free PMC article. Review.
-
Decoding Ubiquitin Modifications by Mass Spectrometry.Adv Exp Med Biol. 2024;1466:1-18. doi: 10.1007/978-981-97-7288-9_1. Adv Exp Med Biol. 2024. PMID: 39546132 Review.
-
Identification of new mechanisms of cellular response to chemotherapy by tracking changes in post-translational modifications by ubiquitin and ubiquitin-like proteins.J Proteome Res. 2014 May 2;13(5):2478-94. doi: 10.1021/pr401258d. Epub 2014 Apr 2. J Proteome Res. 2014. PMID: 24654937
Cited by
-
Pathological implication of protein post-translational modifications in cancer.Mol Aspects Med. 2022 Aug;86:101097. doi: 10.1016/j.mam.2022.101097. Epub 2022 Apr 7. Mol Aspects Med. 2022. PMID: 35400524 Free PMC article. Review.
-
Radiosensitization of Cancer Cells by Inactivation of Cullin-RING E3 Ubiquitin Ligases.Transl Oncol. 2012 Oct;5(5):305-12. doi: 10.1593/tlo.12229. Epub 2012 Oct 1. Transl Oncol. 2012. PMID: 23066438 Free PMC article.
-
Role of protein Post-translational modifications in enterovirus infection.Front Microbiol. 2024 Feb 26;15:1341599. doi: 10.3389/fmicb.2024.1341599. eCollection 2024. Front Microbiol. 2024. PMID: 38596371 Free PMC article. Review.
References
-
- Eng Jimmy K, M A L, Yates John R III. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. JASMS. 1994;5:976–989. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

