Prediction of dinucleotide-specific RNA-binding sites in proteins
- PMID: 22373260
- PMCID: PMC3278845
- DOI: 10.1186/1471-2105-12-S13-S5
Prediction of dinucleotide-specific RNA-binding sites in proteins
Abstract
Background: Regulation of gene expression, protein synthesis, replication and assembly of many viruses involve RNA-protein interactions. Although some successful computational tools have been reported to recognize RNA binding sites in proteins, the problem of specificity remains poorly investigated. After the nucleotide base composition, the dinucleotide is the smallest unit of RNA sequence information and many RNA-binding proteins simply bind to regions enriched in one dinucleotide. Interaction preferences of protein subsequences and dinucleotides can be inferred from protein-RNA complex structures, enabling a training-based prediction approach.
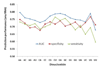
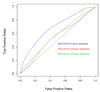
Results: We analyzed basic statistics of amino acid-dinucleotide contacts in protein-RNA complexes and found their pairing preferences could be identified. Using a standard approach to represent protein subsequences by their evolutionary profile, we trained neural networks to predict multiclass target vectors corresponding to 16 possible contacting dinucleotide subsequences. In the cross-validation experiments, the accuracies of the optimum network, measured as areas under the curve (AUC) of the receiver operating characteristic (ROC) graphs, were in the range of 65-80%.
Conclusions: Dinucleotide-specific contact predictions have also been extended to the prediction of interacting protein and RNA fragment pairs, which shows the applicability of this method to predict targets of RNA-binding proteins. A web server predicting the 16-dimensional contact probability matrix directly from a user-defined protein sequence was implemented and made available at: http://tardis.nibio.go.jp/netasa/srcpred.
Figures



Similar articles
-
Prediction of mono- and di-nucleotide-specific DNA-binding sites in proteins using neural networks.BMC Struct Biol. 2009 May 13;9:30. doi: 10.1186/1472-6807-9-30. BMC Struct Biol. 2009. PMID: 19439068 Free PMC article.
-
Prediction of RNA-binding amino acids from protein and RNA sequences.BMC Bioinformatics. 2011;12 Suppl 13(Suppl 13):S7. doi: 10.1186/1471-2105-12-S13-S7. Epub 2011 Nov 30. BMC Bioinformatics. 2011. PMID: 22373313 Free PMC article.
-
Prediction of RNA-binding residues in proteins from primary sequence using an enriched random forest model with a novel hybrid feature.Proteins. 2011 Apr;79(4):1230-9. doi: 10.1002/prot.22958. Epub 2011 Jan 25. Proteins. 2011. PMID: 21268114
-
Computational Prediction of RNA-Binding Proteins and Binding Sites.Int J Mol Sci. 2015 Nov 3;16(11):26303-17. doi: 10.3390/ijms161125952. Int J Mol Sci. 2015. PMID: 26540053 Free PMC article. Review.
-
Computational methods for prediction of protein-RNA interactions.J Struct Biol. 2012 Sep;179(3):261-8. doi: 10.1016/j.jsb.2011.10.001. Epub 2011 Oct 12. J Struct Biol. 2012. PMID: 22019768 Review.
Cited by
-
Systematic Analysis of the Binding Surfaces between tRNAs and Their Respective Aminoacyl tRNA Synthetase Based on Structural and Evolutionary Data.Front Genet. 2018 Jan 8;8:227. doi: 10.3389/fgene.2017.00227. eCollection 2017. Front Genet. 2018. PMID: 29358943 Free PMC article.
-
Twenty years of advances in prediction of nucleic acid-binding residues in protein sequences.Brief Bioinform. 2024 Nov 22;26(1):bbaf016. doi: 10.1093/bib/bbaf016. Brief Bioinform. 2024. PMID: 39833102 Free PMC article. Review.
-
Comprehensive Survey and Comparative Assessment of RNA-Binding Residue Predictions with Analysis by RNA Type.Int J Mol Sci. 2020 Sep 19;21(18):6879. doi: 10.3390/ijms21186879. Int J Mol Sci. 2020. PMID: 32961749 Free PMC article. Review.
-
Deciphering RNA-Recognition Patterns of Intrinsically Disordered Proteins.Int J Mol Sci. 2018 May 29;19(6):1595. doi: 10.3390/ijms19061595. Int J Mol Sci. 2018. PMID: 29843482 Free PMC article.
-
DRNApred, fast sequence-based method that accurately predicts and discriminates DNA- and RNA-binding residues.Nucleic Acids Res. 2017 Jun 2;45(10):e84. doi: 10.1093/nar/gkx059. Nucleic Acids Res. 2017. PMID: 28132027 Free PMC article.
References
-
- Zheng S, Robertson T, Varani G. A knowledge-based potential function predicts the specificity and relative binding energy of RNA-binding proteins. FEBS J. 2007;274:6378–6391. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

