User centered and ontology based information retrieval system for life sciences
- PMID: 22373375
- PMCID: PMC3434427
- DOI: 10.1186/1471-2105-13-S1-S4
User centered and ontology based information retrieval system for life sciences
Abstract
Background: Because of the increasing number of electronic resources, designing efficient tools to retrieve and exploit them is a major challenge. Some improvements have been offered by semantic Web technologies and applications based on domain ontologies. In life science, for instance, the Gene Ontology is widely exploited in genomic applications and the Medical Subject Headings is the basis of biomedical publications indexation and information retrieval process proposed by PubMed. However current search engines suffer from two main drawbacks: there is limited user interaction with the list of retrieved resources and no explanation for their adequacy to the query is provided. Users may thus be confused by the selection and have no idea on how to adapt their queries so that the results match their expectations.
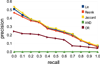
Results: This paper describes an information retrieval system that relies on domain ontology to widen the set of relevant documents that is retrieved and that uses a graphical rendering of query results to favor user interactions. Semantic proximities between ontology concepts and aggregating models are used to assess documents adequacy with respect to a query. The selection of documents is displayed in a semantic map to provide graphical indications that make explicit to what extent they match the user's query; this man/machine interface favors a more interactive and iterative exploration of data corpus, by facilitating query concepts weighting and visual explanation. We illustrate the benefit of using this information retrieval system on two case studies one of which aiming at collecting human genes related to transcription factors involved in hemopoiesis pathway.
Conclusions: The ontology based information retrieval system described in this paper (OBIRS) is freely available at: http://www.ontotoolkit.mines-ales.fr/ObirsClient/. This environment is a first step towards a user centred application in which the system enlightens relevant information to provide decision help.
Figures


References
-
- Vallet D, Fernandez M, Castells P. Proceedings of the 2nd European Semantic Web Conference (ESWC 2005), Volume 3532 of Lecture Notes in Computer Science. Springer Verlag; 2005. An ontology-based information retrieval model; pp. 103–110.
-
- Peltonen J, Aidos H, Gehlenborg N, Brazma A, Kaski S. An information retrieval perspective on visualization of gene expression data with ontological annotation. IEEE International Conference on Acoustics, Speech, and Signal Processing. 2010. pp. 2178–2181.
-
- Bawden D. The dark side of information: overload, anxiety and other paradoxes and pathologies. Journal of Information Science. 2009;35(2):180–191.
-
- Nelson MR. We have the information you want, but getting it will cost you!: held hostage by information overload. Crossroads - Special issue on the Internet. 1994;1(1):11–15.
-
- Christopher DM, Prabhakar R, Hinrich S. Introduction to Information Retrieval. Cambridge University Press; 2008.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

