Evaluation of a LASSO regression approach on the unrelated samples of Genetic Analysis Workshop 17
- PMID: 22373385
- PMCID: PMC3287844
- DOI: 10.1186/1753-6561-5-S9-S12
Evaluation of a LASSO regression approach on the unrelated samples of Genetic Analysis Workshop 17
Abstract
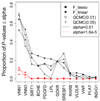
The Genetic Analysis Workshop 17 data we used comprise 697 unrelated individuals genotyped at 24,487 single-nucleotide polymorphisms (SNPs) from a mini-exome scan, using real sequence data for 3,205 genes annotated by the 1000 Genomes Project and simulated phenotypes. We studied 200 sets of simulated phenotypes of trait Q2. An important feature of this data set is that most SNPs are rare, with 87% of the SNPs having a minor allele frequency less than 0.05. For rare SNP detection, in this study we performed a least absolute shrinkage and selection operator (LASSO) regression and F tests at the gene level and calculated the generalized degrees of freedom to avoid any selection bias. For comparison, we also carried out linear regression and the collapsing method, which sums the rare SNPs, modified for a quantitative trait and with two different allele frequency thresholds. The aim of this paper is to evaluate these four approaches in this mini-exome data and compare their performance in terms of power and false positive rates. In most situations the LASSO approach is more powerful than linear regression and collapsing methods. We also note the difficulty in determining the optimal threshold for the collapsing method and the significant role that linkage disequilibrium plays in detecting rare causal SNPs. If a rare causal SNP is in strong linkage disequilibrium with a common marker in the same gene, power will be much improved.
Figures

References
LinkOut - more resources
Full Text Sources

