Genes adopt non-optimal codon usage to generate cell cycle-dependent oscillations in protein levels
- PMID: 22373820
- PMCID: PMC3293633
- DOI: 10.1038/msb.2012.3
Genes adopt non-optimal codon usage to generate cell cycle-dependent oscillations in protein levels
Abstract
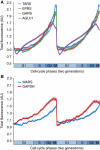
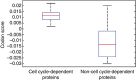
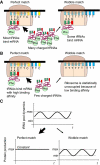
The cell cycle is a temporal program that regulates DNA synthesis and cell division. When we compared the codon usage of cell cycle-regulated genes with that of other genes, we discovered that there is a significant preference for non-optimal codons. Moreover, genes encoding proteins that cycle at the protein level exhibit non-optimal codon preferences. Remarkably, cell cycle-regulated genes expressed in different phases display different codon preferences. Here, we show empirically that transfer RNA (tRNA) expression is indeed highest in the G2 phase of the cell cycle, consistent with the non-optimal codon usage of genes expressed at this time, and lowest toward the end of G1, reflecting the optimal codon usage of G1 genes. Accordingly, protein levels of human glycyl-, threonyl-, and glutamyl-prolyl tRNA synthetases were found to oscillate, peaking in G2/M phase. In light of our findings, we propose that non-optimal (wobbly) matching codons influence protein synthesis during the cell cycle. We describe a new mathematical model that shows how codon usage can give rise to cell-cycle regulation. In summary, our data indicate that cells exploit wobbling to generate cell cycle-dependent dynamics of proteins.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Akashi H, Eyre-Walker A (1998) Translational selection and molecular evolution. Curr Opin Genet Dev 8: 688–693 - PubMed
-
- Cannarozzi G, Schraudolph NN, Faty M, von Rohr P, Friberg MT, Roth AC, Gonnet P, Gonnet G, Barral Y (2010) A role for codon order in translation dynamics. Cell 141: 355–367 - PubMed
-
- Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ, Davis RW (1998) A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell 2: 65–73 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

