Human postmeiotic sex chromatin and its impact on sex chromosome evolution
- PMID: 22375025
- PMCID: PMC3337429
- DOI: 10.1101/gr.135046.111
Human postmeiotic sex chromatin and its impact on sex chromosome evolution
Abstract
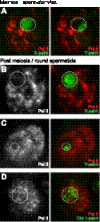
Sex chromosome inactivation is essential epigenetic programming in male germ cells. However, it remains largely unclear how epigenetic silencing of sex chromosomes impacts the evolution of the mammalian genome. Here we demonstrate that male sex chromosome inactivation is highly conserved between humans and mice and has an impact on the genetic evolution of human sex chromosomes. We show that, in humans, sex chromosome inactivation established during meiosis is maintained into spermatids with the silent compartment postmeiotic sex chromatin (PMSC). Human PMSC is illuminated with epigenetic modifications such as trimethylated lysine 9 of histone H3 and heterochromatin proteins CBX1 and CBX3, which implicate a conserved mechanism underlying the maintenance of sex chromosome inactivation in mammals. Furthermore, our analyses suggest that male sex chromosome inactivation has impacted multiple aspects of the evolutionary history of mammalian sex chromosomes: amplification of copy number, retrotranspositions, acquisition of de novo genes, and acquisition of different expression profiles. Most strikingly, profiles of escape genes from postmeiotic silencing diverge significantly between humans and mice. Escape genes exhibit higher rates of amino acid changes compared with non-escape genes, suggesting that they are beneficial for reproductive fitness and may allow mammals to cope with conserved postmeiotic silencing during the evolutionary past. Taken together, we propose that the epigenetic silencing mechanism impacts the genetic evolution of sex chromosomes and contributed to speciation and reproductive diversity in mammals.
Figures



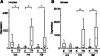

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
