A unique chromosomal rearrangement in the Cryptococcus neoformans var. grubii type strain enhances key phenotypes associated with virulence
- PMID: 22375073
- PMCID: PMC3302566
- DOI: 10.1128/mBio.00310-11
A unique chromosomal rearrangement in the Cryptococcus neoformans var. grubii type strain enhances key phenotypes associated with virulence
Abstract
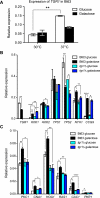
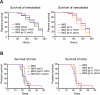
The accumulation of genomic structural variation between closely related populations over time can lead to reproductive isolation and speciation. The fungal pathogen Cryptococcus is thought to have recently diversified, forming a species complex containing members with distinct morphologies, distributions, and pathologies of infection. We have investigated structural changes in genomic architecture such as inversions and translocations that distinguish the most pathogenic variety, Cryptococcus neoformans var. grubii, from the less clinically prevalent Cryptococcus neoformans var. neoformans and Cryptococcus gattii. Synteny analysis between the genomes of the three Cryptococcus species/varieties (strains H99, JEC21, and R265) reveals that C. neoformans var. grubii possesses surprisingly few unique genomic rearrangements. All but one are relatively small and are shared by all molecular subtypes of C. neoformans var. grubii. In contrast, the large translocation peculiar to the C. neoformans var. grubii type strain is found in all tested subcultures from multiple laboratories, suggesting that it has possessed this rearrangement since its isolation from a human clinical sample. Furthermore, we find that the translocation directly disrupts two genes. The first of these encodes a novel protein involved in metabolism of glucose at human body temperature and affects intracellular levels of trehalose. The second encodes a homeodomain-containing transcription factor that modulates melanin production. Both mutations would be predicted to increase pathogenicity; however, when recreated in an alternate genetic background, these mutations do not affect virulence in animal models. The type strain of C. neoformans var. grubii in which the majority of molecular studies have been performed is therefore atypical for carbon metabolism and key virulence attributes.
Importance: The fungal pathogen Cryptococcus is a major cause of mortality among the immunocompromised population, primarily in AIDS patients of sub-Saharan Africa. Most research into the particular variety of Cryptococcus responsible for the vast majority of infections, Cryptococcus neoformans var. grubii, is performed using the type strain isolated in 1978 from a Hodgkin's disease patient from North Carolina. We have determined that this particular isolate contains a chromosomal translocation that directly interrupts two genes, which all descendants of this strain from various research laboratories appear to possess. Disruption of these two genes affects multiple virulence factors of Cryptococcus, particularly the ability to grow at human body temperature, which could have wide-ranging implications for molecular genetic studies and virulence assays using this important strain.
Figures




References
-
- Rieseberg LH. 2001. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16:351–358 - PubMed
-
- Delneri D, et al. 2003. Engineering evolution to study speciation in yeasts. Nature 422:68–72 - PubMed
-
- Navarro A, Barton NH. 2003. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution 57:447–459 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

