Alu-mediated deletion of SOX10 regulatory elements in Waardenburg syndrome type 4
- PMID: 22378281
- PMCID: PMC3421117
- DOI: 10.1038/ejhg.2012.29
Alu-mediated deletion of SOX10 regulatory elements in Waardenburg syndrome type 4
Abstract
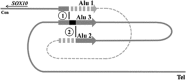
Waardenburg syndrome type 4 (WS4) is a rare neural crest disorder defined by the combination of Waardenburg syndrome (sensorineural hearing loss and pigmentation defects) and Hirschsprung disease (intestinal aganglionosis). Three genes are known to be involved in this syndrome, that is, EDN3 (endothelin-3), EDNRB (endothelin receptor type B), and SOX10. However, 15-35% of WS4 remains unexplained at the molecular level, suggesting that other genes could be involved and/or that mutations within known genes may have escaped previous screenings. Here, we searched for deletions within recently identified SOX10 regulatory sequences and describe the first characterization of a WS4 patient presenting with a large deletion encompassing three of these enhancers. Analysis of the breakpoint region suggests a complex rearrangement involving three Alu sequences that could be mediated by a FosTes/MMBIR replication mechanism. Taken together with recent reports, our results demonstrate that the disruption of highly conserved non-coding elements located within or at a long distance from the coding sequences of key genes can result in several neurocristopathies. This opens up new routes to the molecular dissection of neural crest disorders.
Figures


References
-
- Amiel J, Sproat-Emison E, Garcia-Barcelo M, et al. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. 2008;45:1–14. - PubMed
-
- Pingault V, Ente D, Dastot-Le Moal F, Goossens M, Marlin S, Bondurand N. Review and update of mutations causing Waardenburg syndrome. Hum Mutat. 2010;31:391–406. - PubMed
-
- Inoue K, Khajavi M, Ohyama T, et al. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat Genet. 2004;36:361–369. - PubMed
-
- Amiel J, Benko S, Gordon CT, Lyonnet S. Disruption of long-distance highly conserved noncoding elements in neurocristopathies. Ann NY Acad Sci. 2010;1214:34–46. - PubMed
-
- Emison ES, McCallion AS, Kashuk CS, et al. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–863. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

