Helper virus-independent transcription and multimerization of a satellite RNA associated with cucumber mosaic virus
- PMID: 22379080
- PMCID: PMC3347370
- DOI: 10.1128/JVI.00018-12
Helper virus-independent transcription and multimerization of a satellite RNA associated with cucumber mosaic virus
Abstract
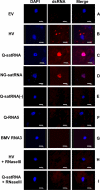
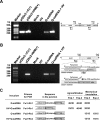
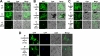
Satellite RNAs are the smallest infectious agents whose replication is thought to be completely dependent on their helper virus (HV). Here we report that, when expressed autonomously in the absence of HV, a variant of satellite RNA (satRNA) associated with Cucumber mosaic virus strain Q (Q-satRNA) has a propensity to localize in the nucleus and be transcribed, generating genomic and antigenomic multimeric forms. The involvement of the nuclear phase of Q-satRNA was further confirmed by confocal microscopy employing in vivo RNA-tagging and double-stranded-RNA-labeling assays. Sequence analyses revealed that the Q-satRNA multimers formed in the absence of HV, compared to when HV is present, are distinguished by the addition of a template-independent heptanucleotide motif at the monomer junctions within the multimers. Collectively, the involvement of a nuclear phase in the replication cycle of Q-satRNA not only provides a valid explanation for its persistent survival in the absence of HV but also suggests a possible evolutionary relationship to viroids that replicate in the nucleus.
Figures


Similar articles
-
A bromodomain-containing host protein mediates the nuclear importation of a satellite RNA of Cucumber mosaic virus.J Virol. 2014 Feb;88(4):1890-6. doi: 10.1128/JVI.03082-13. Epub 2013 Nov 27. J Virol. 2014. PMID: 24284314 Free PMC article.
-
Functional significance of a hepta nucleotide motif present at the junction of Cucumber mosaic virus satellite RNA multimers in helper-virus dependent replication.Virology. 2013 Jan 20;435(2):214-9. doi: 10.1016/j.virol.2012.10.031. Epub 2012 Nov 10. Virology. 2013. PMID: 23146208
-
The non-template functions of helper virus RNAs create optimal replication conditions to enhance the proliferation of satellite RNAs.PLoS Pathog. 2024 Apr 17;20(4):e1012174. doi: 10.1371/journal.ppat.1012174. eCollection 2024 Apr. PLoS Pathog. 2024. PMID: 38630801 Free PMC article.
-
Structure and functional relationships of satellite RNAs of cucumber mosaic virus.Curr Top Microbiol Immunol. 1999;239:37-63. doi: 10.1007/978-3-662-09796-0_3. Curr Top Microbiol Immunol. 1999. PMID: 9893368 Review. No abstract available.
-
Satellite RNAs and Satellite Viruses.Mol Plant Microbe Interact. 2016 Mar;29(3):181-6. doi: 10.1094/MPMI-10-15-0232-FI. Epub 2016 Feb 3. Mol Plant Microbe Interact. 2016. PMID: 26551994 Review.
Cited by
-
Molecular interactions of plant viral satellites.Virus Genes. 2021 Feb;57(1):1-22. doi: 10.1007/s11262-020-01806-9. Epub 2020 Nov 23. Virus Genes. 2021. PMID: 33226576 Review.
-
Rapid evolution of in vivo-selected sequences and structures replacing 20% of a subviral RNA.Virology. 2015 Sep;483:149-62. doi: 10.1016/j.virol.2015.04.002. Epub 2015 May 15. Virology. 2015. PMID: 25974866 Free PMC article.
-
Riboproteomics: A versatile approach for the identification of host protein interaction network in plant pathogenic noncoding RNAs.PLoS One. 2017 Oct 26;12(10):e0186703. doi: 10.1371/journal.pone.0186703. eCollection 2017. PLoS One. 2017. PMID: 29073276 Free PMC article.
-
GAPDH--a recruits a plant virus movement protein to cortical virus replication complexes to facilitate viral cell-to-cell movement.PLoS Pathog. 2014 Nov 20;10(11):e1004505. doi: 10.1371/journal.ppat.1004505. eCollection 2014 Nov. PLoS Pathog. 2014. PMID: 25411849 Free PMC article.
-
Mutations in the capsid protein of Brome mosaic virus affecting encapsidation eliminate vesicle induction in planta: implications for virus cell-to-cell spread.J Virol. 2013 Aug;87(16):8982-92. doi: 10.1128/JVI.01253-13. Epub 2013 Jun 5. J Virol. 2013. PMID: 23741003 Free PMC article.
References
-
- Abraitiene A, Zhao Y, Hammond R. 2008. Nuclear targeting by fragmentation of the potato spindle tuber viroid genome. Biochem. Biophys. Res. Commun. 368:470–475 - PubMed
-
- Albariño CG, Price BD, Eckerle LD, Ball LA. 2001. Characterization and template properties of RNA dimers generated during flock house virus RNA replication. Virology 289:269–282 - PubMed
-
- Annamalai P, Rao AL. 2006. Delivery and expression of functional viral RNA genomes in planta by agroinfiltration. Curr. Protoc. Microbiol. 16:2.1–2.15 - PubMed
-
- Annamalai P, Rao AL. 2005. Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology 338:96–111 - PubMed
-
- Bamunusinghe D, et al. 2009. Analysis of potato virus X replicase and TGBp3 subcellular locations. Virology 393:272–285 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

