The Drosophila 7SK snRNP and the essential role of dHEXIM in development
- PMID: 22379134
- PMCID: PMC3384314
- DOI: 10.1093/nar/gks191
The Drosophila 7SK snRNP and the essential role of dHEXIM in development
Abstract
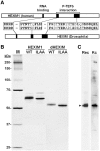
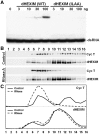
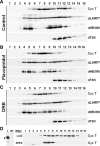
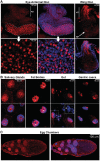
Regulation of the positive transcription elongation factor, P-TEFb, plays a major role in controlling mammalian transcription and this is accomplished in part by controlled release of P-TEFb from the 7SK snRNP that sequesters the kinase in an inactive state. We demonstrate here that a similar P-TEFb control system exists in Drosophila. We show that an RNA previously suggested to be a 7SK homolog is, in fact, associated with P-TEFb, through the action of a homolog of the human HEXIM1/2 proteins (dHEXIM). In addition, a Drosophila La related protein (now called dLARP7) is shown to be the functional homolog of human LARP7. The Drosophila 7SK snRNP (d7SK snRNP) responded to treatment of cells with P-TEFb inhibitors and to nuclease treatment of cell lysates by releasing P-TEFb. Supporting a critical role for the d7SK snRNP in Drosophila development, dLARP7 and dHEXIM were found to be ubiquitously expressed throughout embryos and tissues at all stages. Importantly, knockdown of dHEXIM was embryonic lethal, and reduction of dHEXIM in specific tissues led to serious developmental defects. Our results suggest that regulation of P-TEFb by the d7SK snRNP is essential for the growth and differentiation of tissues required during Drosophila development.
Figures



Similar articles
-
The mechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes a conformational change in 7SK.PLoS One. 2010 Aug 23;5(8):e12335. doi: 10.1371/journal.pone.0012335. PLoS One. 2010. PMID: 20808803 Free PMC article.
-
LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated.Nucleic Acids Res. 2008 Apr;36(7):2219-29. doi: 10.1093/nar/gkn061. Epub 2008 Feb 16. Nucleic Acids Res. 2008. PMID: 18281698 Free PMC article.
-
Release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein (snRNP) activates hexamethylene bisacetamide-inducible protein (HEXIM1) transcription.J Biol Chem. 2014 Apr 4;289(14):9918-25. doi: 10.1074/jbc.M113.539015. Epub 2014 Feb 10. J Biol Chem. 2014. PMID: 24515107 Free PMC article.
-
Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb.Nucleic Acids Res. 2016 Sep 19;44(16):7527-39. doi: 10.1093/nar/gkw585. Epub 2016 Jul 1. Nucleic Acids Res. 2016. PMID: 27369380 Free PMC article. Review.
-
The 7SK snRNP complex: a critical regulator in carcinogenesis.Biochimie. 2025 May 12:S0300-9084(25)00084-7. doi: 10.1016/j.biochi.2025.05.003. Online ahead of print. Biochimie. 2025. PMID: 40368082 Review.
Cited by
-
An evolutionary conserved Hexim1 peptide binds to the Cdk9 catalytic site to inhibit P-TEFb.Proc Natl Acad Sci U S A. 2016 Nov 8;113(45):12721-12726. doi: 10.1073/pnas.1612331113. Epub 2016 Oct 25. Proc Natl Acad Sci U S A. 2016. PMID: 27791144 Free PMC article.
-
Assembly and Exploration of a Single Cell Atlas of the Drosophila Larval Ventral Cord. Identification of Rare Cell Types.Curr Protoc. 2021 Feb;1(2):e37. doi: 10.1002/cpz1.37. Curr Protoc. 2021. PMID: 33600085 Free PMC article.
-
The Bin3 RNA methyltransferase targets 7SK RNA to control transcription and translation.Wiley Interdiscip Rev RNA. 2012 Sep-Oct;3(5):633-47. doi: 10.1002/wrna.1123. Epub 2012 Jun 27. Wiley Interdiscip Rev RNA. 2012. PMID: 22740346 Free PMC article. Review.
-
Transcriptome-wide analysis of pseudouridylation in Drosophila melanogaster.G3 (Bethesda). 2023 Mar 9;13(3):jkac333. doi: 10.1093/g3journal/jkac333. G3 (Bethesda). 2023. PMID: 36534986 Free PMC article.
-
Solution structure of the 5'-terminal hairpin of the 7SK small nuclear RNA.RNA. 2016 Dec;22(12):1844-1858. doi: 10.1261/rna.056523.116. Epub 2016 Oct 20. RNA. 2016. PMID: 27852926 Free PMC article.
References
-
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. - PubMed
-
- Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 1995;270:12335–12338. - PubMed
-
- Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 1996;271:27176–27183. - PubMed
-
- Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell. 2006;21:227–237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

