The BDNF Val66Met polymorphism modulates sleep intensity: EEG frequency- and state-specificity
- PMID: 22379239
- PMCID: PMC3274334
- DOI: 10.5665/sleep.1690
The BDNF Val66Met polymorphism modulates sleep intensity: EEG frequency- and state-specificity
Abstract
Study objectives: EEG slow waves are the hallmark of deep NREM sleep and may reflect the restorative functions of sleep. Evidence suggests that increased sleep slow waves after sleep deprivation reflect plastic synaptic processes, and that brain-derived neurotrophic factor (BDNF) is causally involved in their homeostatic regulation. The functional Val66Met polymorphism of the gene encoding pro-BDNF causes impaired activity-dependent secretion of mature BDNF protein. We investigated whether this polymorphism contributes to the pronounced inter-individual variation in sleep slow wave activity (SWA) in humans.
Setting: Sleep laboratory in temporal isolation unit.
Participants: Eleven heterozygous Met allele carriers and 11 individually sex- and age-matched Val/Val homozygotes.
Interventions: Forty hours prolonged wakefulness.
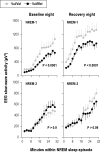
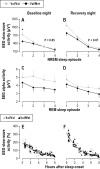
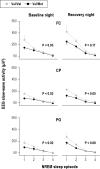
Measurements and results: Cognitive performance, subjective state, and waking and sleep EEG in baseline and after sleep deprivation were studied. Val/Val homozygotes showed better response accuracy than Met allele carriers on a verbal 2-back working memory task. This difference did not reflect genotype-dependent differences in sleepiness, well-being, or sustained attention. In baseline and recovery nights, deep stage 4 sleep and NREM sleep intensity as quantified by EEG SWA (0.75-4.5 Hz) were higher in Val/Val compared to Val/Met genotype. Similar to sleep deprivation, the difference was most pronounced in the first NREM sleep episode. By contrast, increased activity in higher EEG frequencies (> 6 Hz) in wakefulness and REM sleep was distinct from the effects of prolonged wakefulness.
Conclusion: BDNF contributes to the regulation of sleep slow wave oscillations, suggesting that genetically determined variation in neuronal plasticity modulates NREM sleep intensity in humans.
Keywords: Brain derived neurotrophic factor; electroencephalogram; sleep homeostasis; slow wave sleep; synaptic plasticity.
Figures


Comment in
-
Determinants of cortical synchrony.Sleep. 2012 Mar 1;35(3):309-10. doi: 10.5665/sleep.1680. Sleep. 2012. PMID: 22379234 Free PMC article. No abstract available.
References
-
- Achermann P, Borbély AA. Sleep homeostasis and models of sleep regulation. In: Kryger MH, Roth T, Dement WC, editors. Principles and practice of sleep medicine. 5th ed. St. Louis, MI: Elsevier Saunders; 2011. pp. 431–44.
-
- Tononi G, Cirelli C. Sleep function and synaptic homeostasis. Sleep Med Rev. 2006;10:49–62. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

