Conformational analysis of therapeutic proteins by hydroxyl radical protein footprinting
- PMID: 22382679
- PMCID: PMC3326162
- DOI: 10.1208/s12248-012-9336-7
Conformational analysis of therapeutic proteins by hydroxyl radical protein footprinting
Abstract
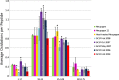
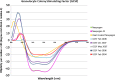
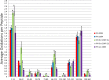
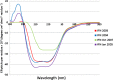
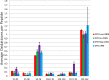
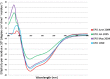
Unlike small molecule drugs, therapeutic protein pharmaceuticals must not only have the correct amino acid sequence and modifications, but also the correct conformation to ensure safety and efficacy. Here, we describe a method for comparison of therapeutic protein conformations by hydroxyl radical protein footprinting using liquid chromatography-mass spectrometry (LC-MS) as an analytical platform. Hydroxyl radical protein footprinting allows for rapid analysis of the conformation of therapeutic proteins based on the apparent rate of oxidation of various amino acids by hydroxyl radicals generated in situ. Conformations of Neupogen®, a patented granulocyte colony-stimulating factor (GCSF), were compared to several expired samples of recombinant GCSF, as well as heat-treated Neupogen®. Conformations of different samples of the therapeutic proteins interferon α-2A and erythropoietin were also compared. Differences in the hydroxyl radical footprint were measured between Neupogen® and the expired or mishandled GCSF samples, and confirmed by circular dichroism spectroscopy. Samples that had identical circular dichroism spectra were also found to be indistinguishable by hydroxyl radical footprinting. The method is applicable to a wide variety of therapeutic proteins and formulations through the use of separations techniques to clean up the protein samples after radical oxidation. The reaction products are stable, allowing for flexibility in sample handling, as well as archiving and reanalysis of samples. Initial screening can be performed on small amounts of therapeutic protein with minimal training in LC-MS, but samples with structural differences from the reference can be more carefully analyzed by LC-MS/MS to attain higher spatial resolution, which can aid in engineering and troubleshooting.
Figures
References
-
- Locatelli F, Del Vecchio L, Pozzoni P. Pure Red-cell aplasia “epidemic”—mystery completely revealed? Perit Dial Int. 2007;27(Supplement 2):S303–S307. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

