Mapping the inputs, analyses, and outputs of biobank research systems to identify sources of incidental findings and individual research results for potential return to participants
- PMID: 22382801
- PMCID: PMC3754832
- DOI: 10.1038/gim.2011.69
Mapping the inputs, analyses, and outputs of biobank research systems to identify sources of incidental findings and individual research results for potential return to participants
Abstract
Progress in the debate over returning incidental findings (IFs) and individual research results (IRRs) to research participants who provide specimens and data to biobanks in genetic and genomic research requires a new tool to allow comparison across heterogeneous biobank research systems and in-depth analysis of the sources and types of findings generated for potential return. This article presents a new visual mapping tool to allow systematic and standardized depiction of (i) the specimens initially collected, (ii) the materials and data sets then created, (iii) the analyses then performed, and finally (iv) the genetic and genomic results generated, including potential IFs and IRRs. For any individual biobank research system, this sequence of four maps can be created to anticipate the sources and types of IFs and IRRs to be generated, to plan how to handle them, and then to manage them responsibly over time. We discuss how this four-map tool was created and describe its application to four national biobank systems, thereby demonstrating that this tool can provide a common platform to visualize biobank content, anticipate how IFs and IRRs will arise in a biobank research context, and inform policy development.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures

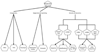

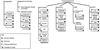
References
-
- Kaufman D, Murphy J, Scott J, Hudson K. Subjects matter: A survey of public opinions about a large genetic cohort study. Genet Med. 2008;10:831–839. - PubMed
-
- Wendler D, Emanuel E. The debate over research on stored biological samples: What do sources think? Arch Intern Med. 2002;162:1457–1462. - PubMed
-
- National Bioethics Advisory Commission. Research Involving Human Biological Materials: Ethical Issues and Policy Guidance. Rockville, MD: 1999.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

