Evolution of siglec-11 and siglec-16 genes in hominins
- PMID: 22383531
- PMCID: PMC3408085
- DOI: 10.1093/molbev/mss077
Evolution of siglec-11 and siglec-16 genes in hominins
Abstract
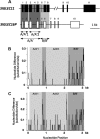
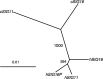
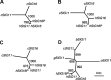
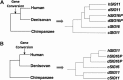
We previously reported a human-specific gene conversion of SIGLEC11 by an adjacent paralogous pseudogene (SIGLEC16P), generating a uniquely human form of the Siglec-11 protein, which is expressed in the human brain. Here, we show that Siglec-11 is expressed exclusively in microglia in all human brains studied-a finding of potential relevance to brain evolution, as microglia modulate neuronal survival, and Siglec-11 recruits SHP-1, a tyrosine phosphatase that modulates microglial biology. Following the recent finding of a functional SIGLEC16 allele in human populations, further analysis of the human SIGLEC11 and SIGLEC16/P sequences revealed an unusual series of gene conversion events between two loci. Two tandem and likely simultaneous gene conversions occurred from SIGLEC16P to SIGLEC11 with a potentially deleterious intervening short segment happening to be excluded. One of the conversion events also changed the 5' untranslated sequence, altering predicted transcription factor binding sites. Both of the gene conversions have been dated to ~1-1.2 Ma, after the emergence of the genus Homo, but prior to the emergence of the common ancestor of Denisovans and modern humans about 800,000 years ago, thus suggesting involvement in later stages of hominin brain evolution. In keeping with this, recombinant soluble Siglec-11 binds ligands in the human brain. We also address a second-round more recent gene conversion from SIGLEC11 to SIGLEC16, with the latter showing an allele frequency of ~0.1-0.3 in a worldwide population study. Initial pseudogenization of SIGLEC16 was estimated to occur at least 3 Ma, which thus preceded the gene conversion of SIGLEC11 by SIGLEC16P. As gene conversion usually disrupts the converted gene, the fact that ORFs of hSIGLEC11 and hSIGLEC16 have been maintained after an unusual series of very complex gene conversion events suggests that these events may have been subject to hominin-specific selection forces.
Figures





Similar articles
-
Possible Influences of Endogenous and Exogenous Ligands on the Evolution of Human Siglecs.Front Immunol. 2018 Dec 4;9:2885. doi: 10.3389/fimmu.2018.02885. eCollection 2018. Front Immunol. 2018. PMID: 30564250 Free PMC article. Review.
-
Coevolution of Siglec-11 and Siglec-16 via gene conversion in primates.BMC Evol Biol. 2017 Nov 23;17(1):228. doi: 10.1186/s12862-017-1075-z. BMC Evol Biol. 2017. PMID: 29169316 Free PMC article.
-
SIGLEC16 encodes a DAP12-associated receptor expressed in macrophages that evolved from its inhibitory counterpart SIGLEC11 and has functional and non-functional alleles in humans.Eur J Immunol. 2008 Aug;38(8):2303-15. doi: 10.1002/eji.200738078. Eur J Immunol. 2008. PMID: 18629938
-
Multiple Genomic Events Altering Hominin SIGLEC Biology and Innate Immunity Predated the Common Ancestor of Humans and Archaic Hominins.Genome Biol Evol. 2020 Jul 1;12(7):1040-1050. doi: 10.1093/gbe/evaa125. Genome Biol Evol. 2020. PMID: 32556248 Free PMC article.
-
Sensing the neuronal glycocalyx by glial sialic acid binding immunoglobulin-like lectins.Neuroscience. 2014 Sep 5;275:113-24. doi: 10.1016/j.neuroscience.2014.05.061. Epub 2014 Jun 9. Neuroscience. 2014. PMID: 24924144 Review.
Cited by
-
Evolution of genetic and genomic features unique to the human lineage.Nat Rev Genet. 2012 Dec;13(12):853-66. doi: 10.1038/nrg3336. Nat Rev Genet. 2012. PMID: 23154808 Free PMC article. Review.
-
Possible Influences of Endogenous and Exogenous Ligands on the Evolution of Human Siglecs.Front Immunol. 2018 Dec 4;9:2885. doi: 10.3389/fimmu.2018.02885. eCollection 2018. Front Immunol. 2018. PMID: 30564250 Free PMC article. Review.
-
Paired Siglec receptors generate opposite inflammatory responses to a human-specific pathogen.EMBO J. 2017 Mar 15;36(6):751-760. doi: 10.15252/embj.201695581. Epub 2017 Jan 18. EMBO J. 2017. PMID: 28100677 Free PMC article.
-
Siglecs as Therapeutic Targets in Cancer.Biology (Basel). 2021 Nov 13;10(11):1178. doi: 10.3390/biology10111178. Biology (Basel). 2021. PMID: 34827170 Free PMC article. Review.
-
Evolution of the exclusively human pathogen Neisseria gonorrhoeae: Human-specific engagement of immunoregulatory Siglecs.Evol Appl. 2019 Jan 3;12(2):337-349. doi: 10.1111/eva.12744. eCollection 2019 Feb. Evol Appl. 2019. PMID: 30697344 Free PMC article.
References
-
- Angata T, Hayakawa T, Yamanaka M, Varki A, Nakamura M. Discovery of Siglec-14, a novel sialic acid receptor undergoing concerted evolution with Siglec-5 in primates. FASEB J. 2006;20:1964–1973. - PubMed
-
- Angata T, Kerr SC, Greaves DR, Varki NM, Crocker PR, Varki A. Cloning and characterization of human Siglec-11. A recently evolved signaling molecule that can interact with SHP-1 and SHP-2 and is expressed by tissue macrophages, including brain microglia. J Biol Chem. 2002;277:24466–24474. - PubMed
-
- Angata T, Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem Rev. 2002;102:439–469. - PubMed
-
- Benatar T, Ratcliffe MJ. Polymorphism of the functional immunoglobulin variable region genes in the chicken by exchange of sequence with donor pseudogenes. Eur J Immunol. 1993;23:2448–2453. - PubMed
-
- Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, Rommens JM. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33:97–101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

