The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes
- PMID: 22383544
- PMCID: PMC3389496
- DOI: 10.1093/molehr/gas013
The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes
Abstract
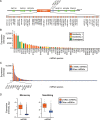
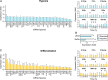
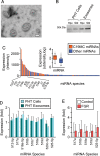
The largest gene cluster of human microRNAs (miRNAs), the chromosome 19 miRNA cluster (C19MC), is exclusively expressed in the placenta and in undifferentiated cells. The precise expression pattern and function of C19MC members are unknown. We sought to profile the relative expression of C19MC miRNAs in primary human trophoblast (PHT) cells and exosomes. Using high-throughput profiling, confirmed by PCR, we found that C19MC miRNAs are among the most abundant miRNAs in term human trophoblasts. Hypoxic stress selectively reduced miR-520c-3p expression at certain time-points with no effect on other C19MC miRNAs. Similarly, differentiation in vitro had a negligible effect on C19MC miRNAs. We found that C19MC miRNAs are the predominant miRNA species expressed in exosomes released from PHT, resembling the profile of trophoblastic cellular miRNA. Predictably, we detected the similar levels of circulating C19MC miRNAs in the serum of healthy pregnant women at term and in women with pregnancies complicated by fetal growth restriction. Our data define the relative expression levels of C19MC miRNAs in trophoblasts and exosomes, and suggest that C19MC miRNAs function in placental-maternal signaling.
Figures
References
-
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. - PMC - PubMed
-
- Atay S, Gercel-Taylor C, Kesimer M, Taylor DD. Morphologic and proteomic characterization of exosomes released by cultured extravillous trophoblast cells. Exp Cell Res. 2011;317:1192–1202. - PubMed
-
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. - PubMed

