Comparison of insertional RNA editing in Myxomycetes
- PMID: 22383871
- PMCID: PMC3285571
- DOI: 10.1371/journal.pcbi.1002400
Comparison of insertional RNA editing in Myxomycetes
Abstract
RNA editing describes the process in which individual or short stretches of nucleotides in a messenger or structural RNA are inserted, deleted, or substituted. A high level of RNA editing has been observed in the mitochondrial genome of Physarum polycephalum. The most frequent editing type in Physarum is the insertion of individual Cs. RNA editing is extremely accurate in Physarum; however, little is known about its mechanism. Here, we demonstrate how analyzing two organisms from the Myxomycetes, namely Physarum polycephalum and Didymium iridis, allows us to test hypotheses about the editing mechanism that can not be tested from a single organism alone. First, we show that using the recently determined full transcriptome information of Physarum dramatically improves the accuracy of computational editing site prediction in Didymium. We use this approach to predict genes in the mitochondrial genome of Didymium and identify six new edited genes as well as one new gene that appears unedited. Next we investigate sequence conservation in the vicinity of editing sites between the two organisms in order to identify sites that harbor the information for the location of editing sites based on increased conservation. Our results imply that the information contained within only nine or ten nucleotides on either side of the editing site (a distance previously suggested through experiments) is not enough to locate the editing sites. Finally, we show that the codon position bias in C insertional RNA editing of these two organisms is correlated with the selection pressure on the respective genes thereby directly testing an evolutionary theory on the origin of this codon bias. Beyond revealing interesting properties of insertional RNA editing in Myxomycetes, our work suggests possible approaches to be used when finding sequence motifs for any biological process fails.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


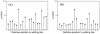
 -values are calculated for shared editing sites in all 16 genes at the (a) first and (b) third codon position. The threshold for statistical significance (
-values are calculated for shared editing sites in all 16 genes at the (a) first and (b) third codon position. The threshold for statistical significance ( as the
as the  -value cut off) is not indicated in the figure as it is far above the top of the graphs.
-value cut off) is not indicated in the figure as it is far above the top of the graphs.
 and
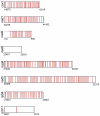
and  are the number of second and third codon position editing sites. Based on the conservation at the second codon position, the genes are separated into (a) four groups and (b) two groups. For the case of 16 known genes, we counted all unambiguous C insertional editing sites in Physarum and Didymium). For the case of 16 known genes + 8 genes in Physarum, we counted all unambiguous C insertional editing sites in Physarum and Didymium for the 16 known genes and unambiguous C insertional editing sites only in Physarum for the additional 8 genes. For 16 known genes + 8 genes in Physarum and Didymium, we counted all unambiguous C insertional editing sites in Physarum and Didymium for all 24 genes.
are the number of second and third codon position editing sites. Based on the conservation at the second codon position, the genes are separated into (a) four groups and (b) two groups. For the case of 16 known genes, we counted all unambiguous C insertional editing sites in Physarum and Didymium). For the case of 16 known genes + 8 genes in Physarum, we counted all unambiguous C insertional editing sites in Physarum and Didymium for the 16 known genes and unambiguous C insertional editing sites only in Physarum for the additional 8 genes. For 16 known genes + 8 genes in Physarum and Didymium, we counted all unambiguous C insertional editing sites in Physarum and Didymium for all 24 genes.
References
-
- Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. - PubMed
-
- Keegan LP, Gallo A, O'Connell MA. The many roles of an RNA editor. Nat Rev Genet. 2001;2:869–878. - PubMed
-
- Horton TL, Landweber LF. Rewriting the information in DNA: RNA editing in kinetoplastids and myxomycetes. Curr Opin Microbiol. 2002;5:620–626. - PubMed
-
- Gott JM, editor. RNA editing. 2007. volume 424 of Methods in Enzymology. Elsevier.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

