Novel transmembrane receptor involved in phagosome transport of lysozymes and β-hexosaminidase in the enteric protozoan Entamoeba histolytica
- PMID: 22383874
- PMCID: PMC3285589
- DOI: 10.1371/journal.ppat.1002539
Novel transmembrane receptor involved in phagosome transport of lysozymes and β-hexosaminidase in the enteric protozoan Entamoeba histolytica
Abstract
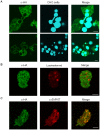
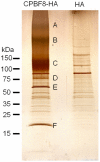
Lysozymes and hexosaminidases are ubiquitous hydrolases in bacteria and eukaryotes. In phagocytic lower eukaryotes and professional phagocytes from higher eukaryotes, they are involved in the degradation of ingested bacteria in phagosomes. In Entamoeba histolytica, which is the intestinal protozoan parasite that causes amoebiasis, phagocytosis plays a pivotal role in the nutrient acquisition and the evasion from the host defense systems. While the content of phagosomes and biochemical and physiological roles of the major phagosomal proteins have been established in E. histolytica, the mechanisms of trafficking of these phagosomal proteins, in general, remain largely unknown. In this study, we identified and characterized for the first time the putative receptor/carrier involved in the transport of the above-mentioned hydrolases to phagosomes. We have shown that the receptor, designated as cysteine protease binding protein family 8 (CPBF8), is localized in lysosomes and mediates transport of lysozymes and β-hexosaminidase α-subunit to phagosomes when the amoeba ingests mammalian cells or Gram-positive bacillus Clostridium perfringens. We have also shown that the binding of CPBF8 to the cargos is mediated by the serine-rich domain, more specifically three serine residues of the domain, which likely contains trifluoroacetic acid-sensitive O-phosphodiester-linked glycan modifications, of CPBF8. We further showed that the repression of CPBF8 by gene silencing reduced the lysozyme and β-hexosaminidase activity in phagosomes and delayed the degradation of C. perfringens. Repression of CPBF8 also resulted in decrease in the cytopathy against the mammalian cells, suggesting that CPBF8 may also be involved in, besides the degradation of ingested bacteria, the pathogenesis against the mammalian hosts. This work represents the first case of the identification of a transport receptor of hydrolytic enzymes responsible for the degradation of microorganisms in phagosomes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
A novel class of cysteine protease receptors that mediate lysosomal transport.Cell Microbiol. 2012 Aug;14(8):1299-317. doi: 10.1111/j.1462-5822.2012.01800.x. Epub 2012 May 14. Cell Microbiol. 2012. PMID: 22486861 Free PMC article.
-
Molecular Dissection of Phagocytosis by Proteomic Analysis in Entamoeba histolytica.Genes (Basel). 2023 Jan 31;14(2):379. doi: 10.3390/genes14020379. Genes (Basel). 2023. PMID: 36833306 Free PMC article. Review.
-
Cysteine protease-binding protein family 6 mediates the trafficking of amylases to phagosomes in the enteric protozoan Entamoeba histolytica.Infect Immun. 2013 May;81(5):1820-9. doi: 10.1128/IAI.00915-12. Epub 2013 Mar 18. Infect Immun. 2013. PMID: 23509141 Free PMC article.
-
Rab7D small GTPase is involved in phago-, trogocytosis and cytoskeletal reorganization in the enteric protozoan Entamoeba histolytica.Cell Microbiol. 2021 Jan;23(1):e13267. doi: 10.1111/cmi.13267. Epub 2020 Nov 1. Cell Microbiol. 2021. PMID: 32975360 Free PMC article.
-
New insights into molecular mechanisms of phagocytosis in Entamoeba histolytica by proteomic analysis.Arch Med Res. 2006 Feb;37(2):244-52. doi: 10.1016/j.arcmed.2005.10.003. Arch Med Res. 2006. PMID: 16380325 Review.
Cited by
-
A novel class of cysteine protease receptors that mediate lysosomal transport.Cell Microbiol. 2012 Aug;14(8):1299-317. doi: 10.1111/j.1462-5822.2012.01800.x. Epub 2012 May 14. Cell Microbiol. 2012. PMID: 22486861 Free PMC article.
-
Immune Response of Amebiasis and Immune Evasion by Entamoeba histolytica.Front Immunol. 2016 May 12;7:175. doi: 10.3389/fimmu.2016.00175. eCollection 2016. Front Immunol. 2016. PMID: 27242782 Free PMC article. Review.
-
Molecular interplays of the Entamoeba histolytica endosomal sorting complexes required for transport during phagocytosis.Front Cell Infect Microbiol. 2022 Oct 27;12:855797. doi: 10.3389/fcimb.2022.855797. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36389174 Free PMC article. Review.
-
Molecular Dissection of Phagocytosis by Proteomic Analysis in Entamoeba histolytica.Genes (Basel). 2023 Jan 31;14(2):379. doi: 10.3390/genes14020379. Genes (Basel). 2023. PMID: 36833306 Free PMC article. Review.
-
Trogocytosis in Unicellular Eukaryotes.Cells. 2021 Nov 1;10(11):2975. doi: 10.3390/cells10112975. Cells. 2021. PMID: 34831198 Free PMC article. Review.
References
-
- Chipman DM, Grisaro V, Sharon N. The binding of oligosaccharides containing N-acetylglucosamine and N-acetylmuramic acid to lysozyme. J Biol Chem. 1967;242:4388–4394. - PubMed
-
- Muraki M, Morikawa M, Jigami Y, Tanaka H. The roles of conserved aromatic amino-acid residues in the active site of human lysozyme: a site-specific mutagenesis study. Biochim Biophys Acta. 1987;916:66–75. - PubMed
-
- Jollès P, Jollès J. What's new in lysozyme research? Always a model system, today and yesterday. Mol Cell Biochem. 1984;53:165–189. - PubMed
-
- Blake CCF, Koenig DF, Mair GA, North ACT, Phillips DC, et al. Structure of hen egg-white lysozyme. A three-dimensional fourier synthesis at 2 angstrom resolution. Nature. 1965;206:757–761. - PubMed
-
- Lemansky P, Hasilik A. Chondroitin sulfate is involved in lysosomal transport of lysozyme in U937 cells. J Cell Sci. 2001;114:345–352. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

