Transient reversal of episome silencing precedes VP16-dependent transcription during reactivation of latent HSV-1 in neurons
- PMID: 22383875
- PMCID: PMC3285597
- DOI: 10.1371/journal.ppat.1002540
Transient reversal of episome silencing precedes VP16-dependent transcription during reactivation of latent HSV-1 in neurons
Abstract
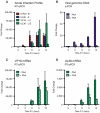
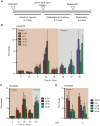
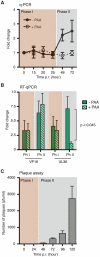
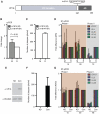
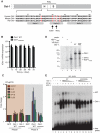
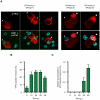
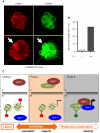
Herpes simplex virus type-1 (HSV-1) establishes latency in peripheral neurons, creating a permanent source of recurrent infections. The latent genome is assembled into chromatin and lytic cycle genes are silenced. Processes that orchestrate reentry into productive replication (reactivation) remain poorly understood. We have used latently infected cultures of primary superior cervical ganglion (SCG) sympathetic neurons to profile viral gene expression following a defined reactivation stimulus. Lytic genes are transcribed in two distinct phases, differing in their reliance on protein synthesis, viral DNA replication and the essential initiator protein VP16. The first phase does not require viral proteins and has the appearance of a transient, widespread de-repression of the previously silent lytic genes. This allows synthesis of viral regulatory proteins including VP16, which accumulate in the cytoplasm of the host neuron. During the second phase, VP16 and its cellular cofactor HCF-1, which is also predominantly cytoplasmic, concentrate in the nucleus where they assemble an activator complex on viral promoters. The transactivation function supplied by VP16 promotes increased viral lytic gene transcription leading to the onset of genome amplification and the production of infectious viral particles. Thus regulated localization of de novo synthesized VP16 is likely to be a critical determinant of HSV-1 reactivation in sympathetic neurons.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Neuronal expression of herpes simplex virus-1 VP16 protein induces pseudorabies virus escape from silencing and reactivation.J Virol. 2024 Jul 23;98(7):e0056124. doi: 10.1128/jvi.00561-24. Epub 2024 Jun 13. J Virol. 2024. PMID: 38869285 Free PMC article.
-
Antagonizing the Glucocorticoid Receptor Impairs Explant-Induced Reactivation in Mice Latently Infected with Herpes Simplex Virus 1.J Virol. 2019 Jun 14;93(13):e00418-19. doi: 10.1128/JVI.00418-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30971470 Free PMC article.
-
De novo synthesis of VP16 coordinates the exit from HSV latency in vivo.PLoS Pathog. 2009 Mar;5(3):e1000352. doi: 10.1371/journal.ppat.1000352. Epub 2009 Mar 27. PLoS Pathog. 2009. PMID: 19325890 Free PMC article.
-
Alphaherpesvirus Latency and Reactivation with a Focus on Herpes Simplex Virus.Curr Issues Mol Biol. 2021;41:267-356. doi: 10.21775/cimb.041.267. Epub 2020 Sep 4. Curr Issues Mol Biol. 2021. PMID: 32883886 Review.
-
Early expression of herpes simplex virus (HSV) proteins and reactivation of latent infection.Folia Microbiol (Praha). 2000;45(1):7-28. doi: 10.1007/BF02817445. Folia Microbiol (Praha). 2000. PMID: 11200675 Review.
Cited by
-
Single-cell transcriptomics identifies Gadd45b as a regulator of herpesvirus-reactivating neurons.EMBO Rep. 2022 Feb 3;23(2):e53543. doi: 10.15252/embr.202153543. Epub 2021 Nov 29. EMBO Rep. 2022. PMID: 34842321 Free PMC article.
-
Molecular Aspects of Varicella-Zoster Virus Latency.Viruses. 2018 Jun 28;10(7):349. doi: 10.3390/v10070349. Viruses. 2018. PMID: 29958408 Free PMC article. Review.
-
Neuronal miR-9 promotes HSV-1 epigenetic silencing and latency by repressing Oct-1 and Onecut family genes.Nat Commun. 2024 Mar 5;15(1):1991. doi: 10.1038/s41467-024-46057-6. Nat Commun. 2024. PMID: 38443365 Free PMC article.
-
HCF1 and OCT2 Cooperate with EBNA1 To Enhance OriP-Dependent Transcription and Episome Maintenance of Latent Epstein-Barr Virus.J Virol. 2016 May 12;90(11):5353-5367. doi: 10.1128/JVI.00239-16. Print 2016 Jun 1. J Virol. 2016. PMID: 27009953 Free PMC article.
-
Stress Flips a Chromatin Switch to Wake Up Latent Virus.Cell Host Microbe. 2015 Dec 9;18(6):639-41. doi: 10.1016/j.chom.2015.11.011. Cell Host Microbe. 2015. PMID: 26651937 Free PMC article.
References
-
- Knipe DM, Cliffe A. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol. 2008;6:211–221. - PubMed
-
- Stevens JG, Wagner EK, Devi-Rao GB, Cook ML, Feldman LT. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056–1059. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 HD023315/HD/NICHD NIH HHS/United States
- R01 GM056927/GM/NIGMS NIH HHS/United States
- NS21072/NS/NINDS NIH HHS/United States
- R01 NS021072/NS/NINDS NIH HHS/United States
- T32 AI007180/AI/NIAID NIH HHS/United States
- R01 GM061139/GM/NIGMS NIH HHS/United States
- S10RR017970/RR/NCRR NIH HHS/United States
- AI073898/AI/NIAID NIH HHS/United States
- GM056927/GM/NIGMS NIH HHS/United States
- R01 AI073898/AI/NIAID NIH HHS/United States
- R56 NS021072/NS/NINDS NIH HHS/United States
- HD23315/HD/NICHD NIH HHS/United States
- GM61139/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

