Coexpression network analysis in abdominal and gluteal adipose tissue reveals regulatory genetic loci for metabolic syndrome and related phenotypes
- PMID: 22383892
- PMCID: PMC3285582
- DOI: 10.1371/journal.pgen.1002505
Coexpression network analysis in abdominal and gluteal adipose tissue reveals regulatory genetic loci for metabolic syndrome and related phenotypes
Abstract
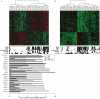
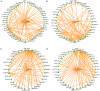
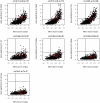
Metabolic Syndrome (MetS) is highly prevalent and has considerable public health impact, but its underlying genetic factors remain elusive. To identify gene networks involved in MetS, we conducted whole-genome expression and genotype profiling on abdominal (ABD) and gluteal (GLU) adipose tissue, and whole blood (WB), from 29 MetS cases and 44 controls. Co-expression network analysis for each tissue independently identified nine, six, and zero MetS-associated modules of coexpressed genes in ABD, GLU, and WB, respectively. Of 8,992 probesets expressed in ABD or GLU, 685 (7.6%) were expressed in ABD and 51 (0.6%) in GLU only. Differential eigengene network analysis of 8,256 shared probesets detected 22 shared modules with high preservation across adipose depots (D(ABD-GLU) = 0.89), seven of which were associated with MetS (FDR P<0.01). The strongest associated module, significantly enriched for immune response-related processes, contained 94/620 (15%) genes with inter-depot differences. In an independent cohort of 145/141 twins with ABD and WB longitudinal expression data, median variability in ABD due to familiality was greater for MetS-associated versus un-associated modules (ABD: 0.48 versus 0.18, P = 0.08; GLU: 0.54 versus 0.20, P = 7.8×10(-4)). Cis-eQTL analysis of probesets associated with MetS (FDR P<0.01) and/or inter-depot differences (FDR P<0.01) provided evidence for 32 eQTLs. Corresponding eSNPs were tested for association with MetS-related phenotypes in two GWAS of >100,000 individuals; rs10282458, affecting expression of RARRES2 (encoding chemerin), was associated with body mass index (BMI) (P = 6.0×10(-4)); and rs2395185, affecting inter-depot differences of HLA-DRB1 expression, was associated with high-density lipoprotein (P = 8.7×10(-4)) and BMI-adjusted waist-to-hip ratio (P = 2.4×10(-4)). Since many genes and their interactions influence complex traits such as MetS, integrated analysis of genotypes and coexpression networks across multiple tissues relevant to clinical traits is an efficient strategy to identify novel associations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

