Sequestration of highly expressed mRNAs in cytoplasmic granules, P-bodies, and stress granules enhances cell viability
- PMID: 22383896
- PMCID: PMC3285586
- DOI: 10.1371/journal.pgen.1002527
Sequestration of highly expressed mRNAs in cytoplasmic granules, P-bodies, and stress granules enhances cell viability
Abstract
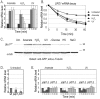
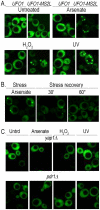
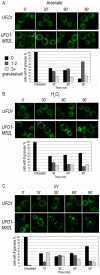
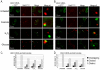
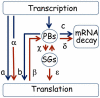
Transcriptome analyses indicate that a core 10%-15% of the yeast genome is modulated by a variety of different stresses. However, not all the induced genes undergo translation, and null mutants of many induced genes do not show elevated sensitivity to the particular stress. Elucidation of the RNA lifecycle reveals accumulation of non-translating mRNAs in cytoplasmic granules, P-bodies, and stress granules for future regulation. P-bodies contain enzymes for mRNA degradation; under stress conditions mRNAs may be transferred to stress granules for storage and return to translation. Protein degradation by the ubiquitin-proteasome system is elevated by stress; and here we analyzed the steady state levels, decay, and subcellular localization of the mRNA of the gene encoding the F-box protein, UFO1, that is induced by stress. Using the MS2L mRNA reporter system UFO1 mRNA was observed in granules that colocalized with P-bodies and stress granules. These P-bodies stored diverse mRNAs. Granules of two mRNAs transported prior to translation, ASH1-MS2L and OXA1-MS2L, docked with P-bodies. HSP12 mRNA that gave rise to highly elevated protein levels was not observed in granules under these stress conditions. ecd3, pat1 double mutants that are defective in P-body formation were sensitive to mRNAs expressed ectopically from strong promoters. These highly expressed mRNAs showed elevated translation compared with wild-type cells, and the viability of the mutants was strongly reduced. ecd3, pat1 mutants also exhibited increased sensitivity to different stresses. Our interpretation is that sequestration of highly expressed mRNAs in P-bodies is essential for viability. Storage of mRNAs for future regulation may contribute to the discrepancy between the steady state levels of many stress-induced mRNAs and their proteins. Sorting of mRNAs for future translation or decay by individual cells could generate potentially different phenotypes in a genetically identical population and enhance its ability to withstand stress.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
mRNA with Mammalian Codon Bias Accumulates in Yeast Mutants with Constitutive Stress Granules.Int J Mol Sci. 2020 Feb 12;21(4):1234. doi: 10.3390/ijms21041234. Int J Mol Sci. 2020. PMID: 32059599 Free PMC article.
-
Granules harboring translationally active mRNAs provide a platform for P-body formation following stress.Cell Rep. 2014 Nov 6;9(3):944-54. doi: 10.1016/j.celrep.2014.09.040. Epub 2014 Oct 23. Cell Rep. 2014. PMID: 25437551 Free PMC article.
-
Relationship of GW/P-bodies with stress granules.Adv Exp Med Biol. 2013;768:197-211. doi: 10.1007/978-1-4614-5107-5_12. Adv Exp Med Biol. 2013. PMID: 23224972 Free PMC article. Review.
-
Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae.J Cell Biol. 2010 May 31;189(5):813-27. doi: 10.1083/jcb.200912019. J Cell Biol. 2010. PMID: 20513766 Free PMC article.
-
P bodies, stress granules, and viral life cycles.Cell Host Microbe. 2008 Apr 17;3(4):206-12. doi: 10.1016/j.chom.2008.03.004. Cell Host Microbe. 2008. PMID: 18407064 Free PMC article. Review.
Cited by
-
Effect of RNA on Morphology and Dynamics of Membraneless Organelles.J Phys Chem B. 2021 May 20;125(19):5035-5044. doi: 10.1021/acs.jpcb.1c02286. Epub 2021 May 10. J Phys Chem B. 2021. PMID: 33969989 Free PMC article.
-
Interplay between Inflammation and Cellular Stress Triggered by Flaviviridae Viruses.Front Microbiol. 2016 Aug 25;7:1233. doi: 10.3389/fmicb.2016.01233. eCollection 2016. Front Microbiol. 2016. PMID: 27610098 Free PMC article. Review.
-
Glucocorticoids enhance chemotherapy-driven stress granule assembly and impair granule dynamics, leading to cell death.J Cell Sci. 2022 Jul 15;135(14):jcs259629. doi: 10.1242/jcs.259629. Epub 2022 Jul 26. J Cell Sci. 2022. PMID: 35713120 Free PMC article.
-
Proteomic Analysis of Dhh1 Complexes Reveals a Role for Hsp40 Chaperone Ydj1 in Yeast P-Body Assembly.G3 (Bethesda). 2015 Sep 21;5(11):2497-511. doi: 10.1534/g3.115.021444. G3 (Bethesda). 2015. PMID: 26392412 Free PMC article.
-
Post-transcriptional regulation during stress.FEMS Yeast Res. 2022 Jun 30;22(1):foac025. doi: 10.1093/femsyr/foac025. FEMS Yeast Res. 2022. PMID: 35561747 Free PMC article. Review.
References
-
- Gasch A, Werner-Washburne M. The genomics of yeast responses to environmental stress and starvation. Funct Integr Genomics. 2002;2:181–192. - PubMed
-
- Temple MD, Perrone GG, Dawes IW. Complex cellular responses to reactive oxygen species. Trends Cell Biol. 2005;15:319–326. - PubMed
-
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

