Mosquitoes LTR retrotransposons: a deeper view into the genomic sequence of Culex quinquefasciatus
- PMID: 22383973
- PMCID: PMC3286476
- DOI: 10.1371/journal.pone.0030770
Mosquitoes LTR retrotransposons: a deeper view into the genomic sequence of Culex quinquefasciatus
Abstract
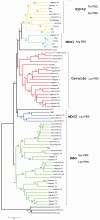
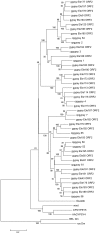
A set of 67 novel LTR-retrotransposon has been identified by in silico analyses of the Culex quinquefasciatus genome using the LTR_STRUC program. The phylogenetic analysis shows that 29 novel and putatively functional LTR-retrotransposons detected belong to the Ty3/gypsy group. Our results demonstrate that, by considering only families containing potentially autonomous LTR-retrotransposons, they account for about 1% of the genome of C. quinquefasciatus. In previous studies it has been estimated that 29% of the genome of C. quinquefasciatus is occupied by mobile genetic elements.The potential role of retrotransposon insertions strictly associated with host genes is described and discussed along with the possible origin of a retrotransposon with peculiar Primer Binding Site region. Finally, we report the presence of a group of 38 retrotransposons, carrying tandem repeated sequences but lacking coding potential, and apparently lacking "master copy" elements from which they could have originated. The features of the repetitive sequences found in these non-autonomous LTR retrotransposons are described, and their possible role discussed.These results integrate the existing data on the genomics of an important virus-borne disease vector.
Conflict of interest statement
Figures

References
-
- Kidwell MG, Lisch DR. Transposable elements as source of genomic variation. In: Craig N, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. American Society for Microbiology Press; 2002. pp. 59–90.
-
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. - PubMed
-
- Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 2008;9:411–412; author reply 414. - PubMed
-
- Finnegan DJ. Transposable elements. Curr Opin Genet Dev. 1992;2:861–867. - PubMed
-
- Deragon JM, Casacuberta JM, Panaud O. Plant transposable elements. Genome Dyn. 2008;4:69–82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

