Investigating the structural impacts of I64T and P311S mutations in APE1-DNA complex: a molecular dynamics approach
- PMID: 22384055
- PMCID: PMC3288039
- DOI: 10.1371/journal.pone.0031677
Investigating the structural impacts of I64T and P311S mutations in APE1-DNA complex: a molecular dynamics approach
Abstract
Background: Elucidating the molecular dynamic behavior of Protein-DNA complex upon mutation is crucial in current genomics. Molecular dynamics approach reveals the changes on incorporation of variants that dictate the structure and function of Protein-DNA complexes. Deleterious mutations in APE1 protein modify the physicochemical property of amino acids that affect the protein stability and dynamic behavior. Further, these mutations disrupt the binding sites and prohibit the protein to form complexes with its interacting DNA.
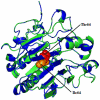
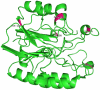
Principal findings: In this study, we developed a rapid and cost-effective method to analyze variants in APE1 gene that are associated with disease susceptibility and evaluated their impacts on APE1-DNA complex dynamic behavior. Initially, two different in silico approaches were used to identify deleterious variants in APE1 gene. Deleterious scores that overlap in these approaches were taken in concern and based on it, two nsSNPs with IDs rs61730854 (I64T) and rs1803120 (P311S) were taken further for structural analysis.
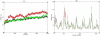
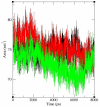
Significance: Different parameters such as RMSD, RMSF, salt bridge, H-bonds and SASA applied in Molecular dynamic study reveals that predicted deleterious variants I64T and P311S alters the structure as well as affect the stability of APE1-DNA interacting functions. This study addresses such new methods for validating functional polymorphisms of human APE1 which is critically involved in causing deficit in repair capacity, which in turn leads to genetic instability and carcinogenesis.
Conflict of interest statement
Figures





References
-
- Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature. 2000;403:451–456. - PubMed
-
- Wilson DM, III, Bohr VA. The Mechanics of base excision repair and its relationship to aging and disease. DNA Repair (Amst.) 2007;6:544–559. - PubMed
-
- Yu E, Gaucher SP, Hadi MZ. Probing conformational changes in Ape1 during the progression of base excision repair. Biochemistry. 2010;49:3786–96. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

