Novel insights into the downstream pathways and targets controlled by transcription factors CREM in the testis
- PMID: 22384077
- PMCID: PMC3285179
- DOI: 10.1371/journal.pone.0031798
Novel insights into the downstream pathways and targets controlled by transcription factors CREM in the testis
Abstract
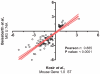
The essential role of the Crem gene in normal sperm development is widely accepted and is confirmed by azoospermia in male mice lacking the Crem gene. The exact number of genes affected by Crem absence is not known, however a large difference has been observed recently between the estimated number of differentially expressed genes found in Crem knock-out (KO) mice compared to the number of gene loci bound by CREM. We therefore re-examined global gene expression in male mice lacking the Crem gene using whole genome transcriptome analysis with Affymetrix microarrays and compared the lists of differentially expressed genes from Crem-/- mice to a dataset of genes where binding of CREM was determined by Chip-seq. We determined the global effect of CREM on spermatogenesis as well as distinguished between primary and secondary effects of the CREM absence. We demonstrated that the absence of Crem deregulates over 4700 genes in KO testis. Among them are 101 genes associated with spermatogenesis 41 of which are bound by CREM and are deregulated in Crem KO testis. Absence of several of these genes in mouse models has proven their importance for normal spermatogenesis and male fertility. Our study showed that the absence of Crem plays a more important role on different aspects of spermatogenesis as estimated previously, with its impact ranging from apoptosis induction to deregulation of major circadian clock genes, steroidogenesis and the cell-cell junction dynamics. Several new genes important for normal spermatogenesis and fertility are down-regulated in KO testis and are therefore possible novel targets of CREM.
Conflict of interest statement
Figures



Similar articles
-
Analysis of CREM-dependent gene expression during mouse spermatogenesis.Mol Cell Endocrinol. 2003 Dec 30;212(1-2):29-39. doi: 10.1016/j.mce.2003.09.023. Mol Cell Endocrinol. 2003. PMID: 14654248
-
Cell-specific occupancy of an extended repertoire of CREM and CREB binding loci in male germ cells.BMC Genomics. 2010 Sep 29;11:530. doi: 10.1186/1471-2164-11-530. BMC Genomics. 2010. PMID: 20920259 Free PMC article.
-
Expression of activator of CREM in the testis (ACT) during normal and impaired spermatogenesis: correlation with CREM expression.Mol Hum Reprod. 2004 Feb;10(2):129-35. doi: 10.1093/molehr/gah012. Mol Hum Reprod. 2004. PMID: 14742698
-
The CREM system in human spermatogenesis.J Endocrinol Invest. 2000 Oct;23(9):578-83. doi: 10.1007/BF03343779. J Endocrinol Invest. 2000. PMID: 11079452 Review.
-
Regulation of gene expression in post-meiotic male germ cells: CREM-signalling pathways and male fertility.Hum Fertil (Camb). 2006 Jun;9(2):73-9. doi: 10.1080/14647270500463400. Hum Fertil (Camb). 2006. PMID: 16825108 Review.
Cited by
-
The glucose-sensing transcription factor MLX balances metabolism and stress to suppress apoptosis and maintain spermatogenesis.PLoS Biol. 2021 Oct 20;19(10):e3001085. doi: 10.1371/journal.pbio.3001085. eCollection 2021 Oct. PLoS Biol. 2021. PMID: 34669700 Free PMC article.
-
TauCstF-64 Mediates Correct mRNA Polyadenylation and Splicing of Activator and Repressor Isoforms of the Cyclic AMP-Responsive Element Modulator (CREM) in Mouse Testis.Biol Reprod. 2016 Feb;94(2):34. doi: 10.1095/biolreprod.115.134684. Epub 2015 Dec 23. Biol Reprod. 2016. PMID: 26700942 Free PMC article.
-
Screening the key microRNAs and transcription factors in prostate cancer based on microRNA functional synergistic relationships.Medicine (Baltimore). 2017 Jan;96(1):e5679. doi: 10.1097/MD.0000000000005679. Medicine (Baltimore). 2017. PMID: 28072703 Free PMC article.
-
Unified single-cell analysis of testis gene regulation and pathology in five mouse strains.Elife. 2019 Jun 25;8:e43966. doi: 10.7554/eLife.43966. Elife. 2019. PMID: 31237565 Free PMC article.
-
An introduction to murine models of atrial fibrillation.Front Physiol. 2012 Aug 3;3:296. doi: 10.3389/fphys.2012.00296. eCollection 2012. Front Physiol. 2012. PMID: 22934047 Free PMC article.
References
-
- Xiao P, Tang A, Yu Z, Gui Y, Cai Z. Gene expression profile of 2058 spermatogenesis-related genes in mice. Biological and Pharmaceutical Bulletin. 2008;31:201–206. - PubMed
-
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biology of Reproduction. 2004;71:319–330. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

