A phylogenetic analysis of the globins in fungi
- PMID: 22384087
- PMCID: PMC3287990
- DOI: 10.1371/journal.pone.0031856
A phylogenetic analysis of the globins in fungi
Abstract
Background: All globins belong to one of three families: the F (flavohemoglobin) and S (sensor) families that exhibit the canonical 3/3 α-helical fold, and the T (truncated 3/3 fold) globins characterized by a shortened 2/2 α-helical fold. All eukaryote 3/3 hemoglobins are related to the bacterial single domain F globins. It is known that Fungi contain flavohemoglobins and single domain S globins. Our aims are to provide a census of fungal globins and to examine their relationships to bacterial globins.
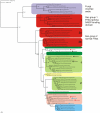
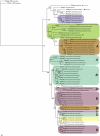
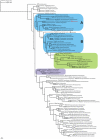
Results: Examination of 165 genomes revealed that globins are present in >90% of Ascomycota and ~60% of Basidiomycota genomes. The S globins occur in Blastocladiomycota and Chytridiomycota in addition to the phyla that have FHbs. Unexpectedly, group 1 T globins were found in one Blastocladiomycota and one Chytridiomycota genome. Phylogenetic analyses were carried out on the fungal globins, alone and aligned with representative bacterial globins. The Saccharomycetes and Sordariomycetes with two FHbs form two widely divergent clusters separated by the remaining fungal sequences. One of the Saccharomycete groups represents a new subfamily of FHbs, comprising a previously unknown N-terminal and a FHb missing the C-terminal moiety of its reductase domain. The two Saccharomycete groups also form two clusters in the presence of bacterial FHbs; the surrounding bacterial sequences are dominated by Proteobacteria and Bacilli (Firmicutes). The remaining fungal FHbs cluster with Proteobacteria and Actinobacteria. The Sgbs cluster separately from their bacterial counterparts, except for the intercalation of two Planctomycetes and a Proteobacterium between the Fungi incertae sedis and the Blastocladiomycota and Chytridiomycota.
Conclusion: Our results are compatible with a model of globin evolution put forward earlier, which proposed that eukaryote F, S and T globins originated via horizontal gene transfer of their bacterial counterparts to the eukaryote ancestor, resulting from the endosymbiotic events responsible for the origin of mitochondria and chloroplasts.
Conflict of interest statement
Figures




References
-
- Keilin D. Occurrence of haemoglobin in yeast and the supposed stabilization of the oxygenated cytochrome oxidase. Nature. 1953;172:390–393. - PubMed
-
- Keilin D, Tissieres A. Haemoglobin in moulds Neuropora crassa and Penicillium notatum. Nature. 1953;172:393–394. - PubMed
-
- Oshino R, Asakura T, Takio K, Oshino N, Chance B, et al. Purification and molecular properties of yeast hemoglobin. Eur J Biochem. 1973;39:581–590. - PubMed
-
- Iwaasa H, Takagi T, Shikama K. Amino acid sequence of yeast hemoglobin. A twodomain structure. J Mol Biol. 1992;227:948–954. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

