3T3 cell lines stably expressing Pax6 or Pax6(5a)--a new tool used for identification of common and isoform specific target genes
- PMID: 22384097
- PMCID: PMC3285655
- DOI: 10.1371/journal.pone.0031915
3T3 cell lines stably expressing Pax6 or Pax6(5a)--a new tool used for identification of common and isoform specific target genes
Abstract
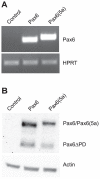
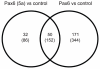
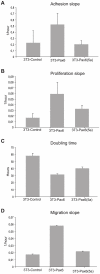
Pax6 and Pax6(5a) are two isoforms of the evolutionary conserved Pax6 gene often co-expressed in specific stochiometric relationship in the brain and the eye during development. The Pax6(5a) protein differs from Pax6 by having a 14 amino acid insert in the paired domain, causing the two proteins to have different DNA binding specificities. Difference in functions during development is proven by the fact that mutations in the 14 amino acid insertion for Pax6(5a) give a slightly different eye phenotype than the one described for Pax6. Whereas quite many Pax6 target genes have been published during the last years, few Pax6(5a) specific target genes have been reported on. However, target genes identified by Pax6 knockout studies can probably be Pax6(5a) targets as well, since this isoform also will be affected by the knockout. In order to identify new Pax6 target genes, and to try to distinguish between genes regulated by Pax6 and Pax6(5a), we generated FlpIn-3T3 cell lines stably expressing Pax6 or Pax6(5a). RNA was harvested from these cell lines and used in gene expression microarrays where we identified a number of genes differentially regulated by Pax6 and Pax6(5a). A majority of these were associated with the extracellular region. By qPCR we verified that Ncam1, Ngef, Sphk1, Dkk3 and Crtap are Pax6(5a) specific target genes, while Tgfbi, Vegfa, EphB2, Klk8 and Edn1 were confirmed as Pax6 specific target genes. Nbl1, Ngfb and seven genes encoding different glycosyl transferases appeared to be regulated by both. Direct binding to the promoters of Crtap, Ctgf, Edn1, Dkk3, Pdgfb and Ngef was verified by ChIP. Furthermore, a change in morphology of the stably transfected Pax6 and Pax6(5a) cells was observed, and the Pax6 expressing cells were shown to have increased proliferation and migration capacities.
Conflict of interest statement
Figures


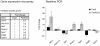


References
-
- Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. - PubMed
-
- Yasuda T, Kajimoto Y, Fujitani Y, Watada H, Yamamoto S, et al. PAX6 mutation as a genetic factor common to aniridia and glucose intolerance. Diabetes. 2002;51:224–230. - PubMed
-
- Mitchell TN, Free SL, Williamson KA, Stevens JM, Churchill AJ, et al. Polymicrogyria and absence of pineal gland due to PAX6 mutation. Ann Neurol. 2003;53:658–663. - PubMed
-
- Estivill-Torrus G, Vitalis T, Fernandez-Llebrez P, Price DJ. The transcription factor Pax6 is required for development of the diencephalic dorsal midline secretory radial glia that form the subcommissural organ. Mech Dev. 2001;109:215–224. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

