Functional desaturase Fads1 (Δ5) and Fads2 (Δ6) orthologues evolved before the origin of jawed vertebrates
- PMID: 22384110
- PMCID: PMC3285190
- DOI: 10.1371/journal.pone.0031950
Functional desaturase Fads1 (Δ5) and Fads2 (Δ6) orthologues evolved before the origin of jawed vertebrates
Abstract
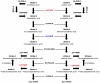
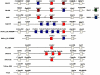
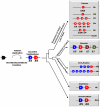
Long-chain polyunsaturated fatty acids (LC-PUFAs) such as arachidonic (ARA), eicosapentaenoic (EPA) and docosahexaenoic (DHA) acids are essential components of biomembranes, particularly in neural tissues. Endogenous synthesis of ARA, EPA and DHA occurs from precursor dietary essential fatty acids such as linoleic and α-linolenic acid through elongation and Δ5 and Δ6 desaturations. With respect to desaturation activities some noteworthy differences have been noted in vertebrate classes. In mammals, the Δ5 activity is allocated to the Fads1 gene, while Fads2 is a Δ6 desaturase. In contrast, teleosts show distinct combinations of desaturase activities (e.g. bifunctional or separate Δ5 and Δ6 desaturases) apparently allocated to Fads2-type genes. To determine the timing of Fads1-Δ5 and Fads2-Δ6 evolution in vertebrates we used a combination of comparative and functional genomics with the analysis of key phylogenetic species. Our data show that Fads1 and Fads2 genes with Δ5 and Δ6 activities respectively, evolved before gnathostome radiation, since the catshark Scyliorhinus canicula has functional orthologues of both gene families. Consequently, the loss of Fads1 in teleosts is a secondary episode, while the existence of Δ5 activities in the same group most likely occurred through independent mutations into Fads2 type genes. Unexpectedly, we also establish that events of Fads1 gene expansion have taken place in birds and reptiles. Finally, a fourth Fads gene (Fads4) was found with an exclusive occurrence in mammalian genomes. Our findings enlighten the history of a crucially important gene family in vertebrate fatty acid metabolism and physiology and provide an explanation of how observed lineage-specific gene duplications, losses and diversifications might be linked to habitat-specific food web structures in different environments and over geological timescales.
Conflict of interest statement
Figures


Similar articles
-
Retention of fatty acyl desaturase 1 (fads1) in Elopomorpha and Cyclostomata provides novel insights into the evolution of long-chain polyunsaturated fatty acid biosynthesis in vertebrates.BMC Evol Biol. 2018 Oct 19;18(1):157. doi: 10.1186/s12862-018-1271-5. BMC Evol Biol. 2018. PMID: 30340454 Free PMC article.
-
Isolation and Functional Characterisation of a fads2 in Rainbow Trout (Oncorhynchus mykiss) with Δ5 Desaturase Activity.PLoS One. 2016 Mar 4;11(3):e0150770. doi: 10.1371/journal.pone.0150770. eCollection 2016. PLoS One. 2016. PMID: 26943160 Free PMC article.
-
A complete enzymatic capacity for long-chain polyunsaturated fatty acid biosynthesis is present in the Amazonian teleost tambaqui, Colossoma macropomum.Comp Biochem Physiol B Biochem Mol Biol. 2019 Jan;227:90-97. doi: 10.1016/j.cbpb.2018.09.003. Epub 2018 Oct 2. Comp Biochem Physiol B Biochem Mol Biol. 2019. PMID: 30290221
-
The impact of fatty acid desaturase genotype on fatty acid status and cardiovascular health in adults.Proc Nutr Soc. 2017 Feb;76(1):64-75. doi: 10.1017/S0029665116000732. Epub 2016 Aug 16. Proc Nutr Soc. 2017. PMID: 27527582 Review.
-
Genetically determined variation in polyunsaturated fatty acid metabolism may result in different dietary requirements.Nestle Nutr Workshop Ser Pediatr Program. 2008;62:35-44; discussion 44-9. doi: 10.1159/000146246. Nestle Nutr Workshop Ser Pediatr Program. 2008. PMID: 18626191 Review.
Cited by
-
Molecular Identification and Functional Characterization of LC-PUFA Biosynthesis Elongase (elovl2) Gene in Chinese Sturgeon (Acipenser sinensis).Animals (Basel). 2024 Aug 14;14(16):2343. doi: 10.3390/ani14162343. Animals (Basel). 2024. PMID: 39199889 Free PMC article.
-
Tecticornia sp. (Samphire)-A Promising Underutilized Australian Indigenous Edible Halophyte.Front Nutr. 2021 Feb 5;8:607799. doi: 10.3389/fnut.2021.607799. eCollection 2021. Front Nutr. 2021. PMID: 33614696 Free PMC article.
-
Solexa-Sequencing Based Transcriptome Study of Plaice Skin Phenotype in Rex Rabbits (Oryctolagus cuniculus).PLoS One. 2015 May 8;10(5):e0124583. doi: 10.1371/journal.pone.0124583. eCollection 2015. PLoS One. 2015. PMID: 25955442 Free PMC article.
-
Dietary Oil Source and Selenium Supplementation Modulate Fads2 and Elovl5 Transcriptional Levels in Liver and Brain of Meagre (Argyrosomus regius).Lipids. 2016 Jun;51(6):729-41. doi: 10.1007/s11745-016-4157-6. Epub 2016 May 12. Lipids. 2016. PMID: 27169705
-
Regulation of Δ6Fads2 Gene Involved in LC-PUFA Biosynthesis Subjected to Fatty Acid in Large Yellow Croaker (Larimichthys crocea) and Rainbow Trout (Oncorhynchus mykiss).Biomolecules. 2022 Apr 30;12(5):659. doi: 10.3390/biom12050659. Biomolecules. 2022. PMID: 35625587 Free PMC article.
References
-
- Tocher DR. Metabolism and functions of lipids and fatty acids in teleost fish. Rev Fisheries Sci. 2003;11:107–184.
-
- Tocher DR. Fatty acid requirements in ontogeny of marine and freshwater fish. Aquaculture Res. 2010;41:717–732.
-
- Burdge GC, Calder PC. Conversion of alpha-linolenic acid to longer-chain polyunsaturated fatty acids in human adults. Reprod Nutr Dev. 2005;45:581–97. - PubMed
-
- Rivers JPW, Sinclair AJ, Crawford MA. Inability of the cat to desaturate essential fatty acids. Nature. 1975;258:171–173. - PubMed
-
- Cook HW, McMaster RCR. Biochemistry of Lipids, Lipoproteins, and Membranes. In: Vance DE, Vance JE, editors. Amsterdam: Elsevier; 2004. pp. 181–204.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

