Cytotoxicity of CD56(bright) NK cells towards autologous activated CD4+ T cells is mediated through NKG2D, LFA-1 and TRAIL and dampened via CD94/NKG2A
- PMID: 22384114
- PMCID: PMC3284517
- DOI: 10.1371/journal.pone.0031959
Cytotoxicity of CD56(bright) NK cells towards autologous activated CD4+ T cells is mediated through NKG2D, LFA-1 and TRAIL and dampened via CD94/NKG2A
Abstract
In mouse models of chronic inflammatory diseases, Natural Killer (NK) cells can play an immunoregulatory role by eliminating chronically activated leukocytes. Indirect evidence suggests that NK cells may also be immunoregulatory in humans. Two subsets of human NK cells can be phenotypically distinguished as CD16(+)CD56(dim) and CD16(dim/-)CD56(bright). An expansion in the CD56(bright) NK cell subset has been associated with clinical responses to therapy in various autoimmune diseases, suggesting an immunoregulatory role for this subset in vivo. Here we compared the regulation of activated human CD4(+) T cells by CD56(dim) and CD56(bright) autologous NK cells in vitro. Both subsets efficiently killed activated, but not resting, CD4(+) T cells. The activating receptor NKG2D, as well as the integrin LFA-1 and the TRAIL pathway, played important roles in this process. Degranulation by NK cells towards activated CD4(+) T cells was enhanced by IL-2, IL-15, IL-12+IL-18 and IFN-α. Interestingly, IL-7 and IL-21 stimulated degranulation by CD56(bright) NK cells but not by CD56(dim) NK cells. NK cell killing of activated CD4(+) T cells was suppressed by HLA-E on CD4(+) T cells, as blocking the interaction between HLA-E and the inhibitory CD94/NKG2A NK cell receptor enhanced NK cell degranulation. This study provides new insight into CD56(dim) and CD56(bright) NK cell-mediated elimination of activated autologous CD4(+) T cells, which potentially may provide an opportunity for therapeutic treatment of chronic inflammation.
Conflict of interest statement
Figures



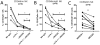
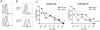
References
-
- Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. - PubMed
-
- Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. - PubMed
-
- Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–244. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

