First dating of a recombination event in mammalian tick-borne flaviviruses
- PMID: 22384119
- PMCID: PMC3285191
- DOI: 10.1371/journal.pone.0031981
First dating of a recombination event in mammalian tick-borne flaviviruses
Abstract
The mammalian tick-borne flavivirus group (MTBFG) contains viruses associated with important human and animal diseases such as encephalitis and hemorrhagic fever. In contrast to mosquito-borne flaviviruses where recombination events are frequent, the evolutionary dynamic within the MTBFG was believed to be essentially clonal. This assumption was challenged with the recent report of several homologous recombinations within the Tick-borne encephalitis virus (TBEV). We performed a thorough analysis of publicly available genomes in this group and found no compelling evidence for the previously identified recombinations. However, our results show for the first time that demonstrable recombination (i.e., with large statistical support and strong phylogenetic evidences) has occurred in the MTBFG, more specifically within the Louping ill virus lineage. Putative parents, recombinant strains and breakpoints were further tested for statistical significance using phylogenetic methods. We investigated the time of divergence between the recombinant and parental strains in a Bayesian framework. The recombination was estimated to have occurred during a window of 282 to 76 years before the present. By unravelling the temporal setting of the event, we adduce hypotheses about the ecological conditions that could account for the observed recombination.
Conflict of interest statement
Figures
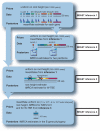
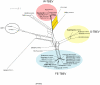

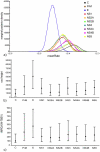
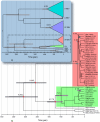
References
-
- Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. - PubMed
-
- Gao GF, Jiang WR, Hussain MH, Venugopal K, Gritsun TS, et al. Sequencing and antigenic studies of a Norwegian virus isolated from encephalomyelitic sheep confirm the existence of louping ill virus outside Great Britain and Ireland. J Gen Virol. 1993;74:109–114. - PubMed
-
- Gao GF, Zanotto PM, Holmes EC, Reid HW, Gould EA. Molecular variation, evolution and geographical distribution of Louping ill virus. Acta Virol. 1997;41:259–268. - PubMed
-
- Grard G, Moureau G, Charrel RN, Lemasson J-J, Gonzalez J-P, et al. Genetic characterization of tick-borne flaviviruses: New insights into evolution, pathogenetic determinants and taxonomy. Virology. 2007;361:80–92. - PubMed
-
- Marin MS, McKenzie J, Gao GF, Reid HW, Antoniadis A, et al. The virus causing encephalomyelitis in sheep in Spain: a new member of the tick-borne encephalitis group. Res Vet Sci. 1995;58:11–13. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

