Contribution of transcription factor binding site motif variants to condition-specific gene expression patterns in budding yeast
- PMID: 22384202
- PMCID: PMC3285675
- DOI: 10.1371/journal.pone.0032274
Contribution of transcription factor binding site motif variants to condition-specific gene expression patterns in budding yeast
Abstract
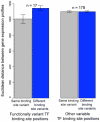
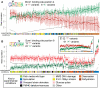
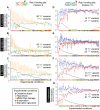
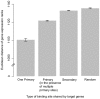
It is now experimentally well known that variant sequences of a cis transcription factor binding site motif can contribute to differential regulation of genes. We characterize the relationship between motif variants and gene expression by analyzing expression microarray data and binding site predictions. To accomplish this, we statistically detect motif variants with effects that differ among environments. Such environmental specificity may be due to either affinity differences between variants or, more likely, differential interactions of TFs bound to these variants with cofactors, and with differential presence of cofactors across environments. We examine conservation of functional variants across four Saccharomyces species, and find that about a third of transcription factors have target genes that are differentially expressed in a condition-specific manner that is correlated with the nucleotide at variant motif positions. We find good correspondence between our results and some cases in the experimental literature (Reb1, Sum1, Mcm1, and Rap1). These results and growing consensus in the literature indicates that motif variants may often be functionally distinct, that this may be observed in genomic data, and that variants play an important role in condition-specific gene regulation.
Conflict of interest statement
Figures

References
-
- Rockman MV, Wray GA. Abundant raw material for cis-regulatory evolution in humans. Mol Biol Evol. 2002;19:1991–2004. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

