Mammalian genes preferentially co-retained in radiation hybrid panels tend to avoid coexpression
- PMID: 22384204
- PMCID: PMC3286474
- DOI: 10.1371/journal.pone.0032284
Mammalian genes preferentially co-retained in radiation hybrid panels tend to avoid coexpression
Abstract
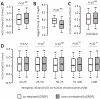
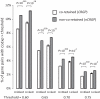
Coexpression has been frequently used to explore modules of functionally related genes in eukaryotic genomes. However, we found that genetically interacting mammalian genes identified through radiation hybrid (RH) genotypes tend not to be coexpressed across tissues. This pattern remained unchanged after controlling for potential confounding factors, including chromosomal linkage, chromosomal distance, and gene duplication. Because >99.9% of the genetically interacting genes were identified according to the higher co-retention frequencies, our observation implies that coexpression is not necessarily an indication of the need for the co-presence of two genes in the genome, which is a prerequisite for cofunctionality of their coding proteins in the cell. Therefore, coexpression information must be applied cautiously to the exploration of the functional relatedness of genes in a genome.
Conflict of interest statement
Figures
Similar articles
-
Construction of a radiation hybrid map of chicken chromosome 2 and alignment to the chicken draft sequence.BMC Genomics. 2005 Feb 4;6:12. doi: 10.1186/1471-2164-6-12. BMC Genomics. 2005. PMID: 15693999 Free PMC article.
-
Chromosome-specific single-locus FISH probes allow anchorage of an 1800-marker integrated radiation-hybrid/linkage map of the domestic dog genome to all chromosomes.Genome Res. 2001 Oct;11(10):1784-95. doi: 10.1101/gr.189401. Genome Res. 2001. PMID: 11591656 Free PMC article.
-
Coexpression of linked genes in Mammalian genomes is generally disadvantageous.Mol Biol Evol. 2008 Aug;25(8):1555-65. doi: 10.1093/molbev/msn101. Epub 2008 Apr 24. Mol Biol Evol. 2008. PMID: 18440951 Free PMC article.
-
[Genome radiation hybrid mapping: summary and future directions].Genetika. 2002 May;38(5):581-94. Genetika. 2002. PMID: 12068541 Review. Russian.
-
Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes.Genomics. 2008 Mar;91(3):243-8. doi: 10.1016/j.ygeno.2007.11.002. Epub 2007 Dec 21. Genomics. 2008. PMID: 18082363 Review.
Cited by
-
Functional characterization of motif sequences under purifying selection.Nucleic Acids Res. 2013 Feb 1;41(4):2105-20. doi: 10.1093/nar/gks1456. Epub 2013 Jan 8. Nucleic Acids Res. 2013. PMID: 23303791 Free PMC article.
References
-
- Hurst LD, Pal C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet. 2004;5:299–310. - PubMed
-
- Cohen BA, Mitra RD, Hughes JD, Church GM. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet. 2000;26:183–186. - PubMed
-
- Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

