The TAL effector PthA4 interacts with nuclear factors involved in RNA-dependent processes including a HMG protein that selectively binds poly(U) RNA
- PMID: 22384209
- PMCID: PMC3285215
- DOI: 10.1371/journal.pone.0032305
The TAL effector PthA4 interacts with nuclear factors involved in RNA-dependent processes including a HMG protein that selectively binds poly(U) RNA
Erratum in
-
Correction: The TAL Effector PthA4 Interacts with Nuclear Factors Involved in RNA-Dependent Processes Including a HMG Protein That Selectively Binds Poly(U) RNA.PLoS One. 2015 Jul 31;10(7):e0134818. doi: 10.1371/journal.pone.0134818. eCollection 2015. PLoS One. 2015. PMID: 26230714 Free PMC article. No abstract available.
Abstract
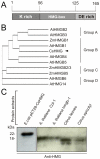
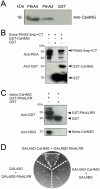
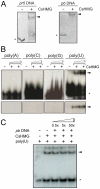
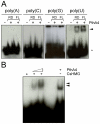
Plant pathogenic bacteria utilize an array of effector proteins to cause disease. Among them, transcriptional activator-like (TAL) effectors are unusual in the sense that they modulate transcription in the host. Although target genes and DNA specificity of TAL effectors have been elucidated, how TAL proteins control host transcription is poorly understood. Previously, we showed that the Xanthomonas citri TAL effectors, PthAs 2 and 3, preferentially targeted a citrus protein complex associated with transcription control and DNA repair. To extend our knowledge on the mode of action of PthAs, we have identified new protein targets of the PthA4 variant, required to elicit canker on citrus. Here we show that all the PthA4-interacting proteins are DNA and/or RNA-binding factors implicated in chromatin remodeling and repair, gene regulation and mRNA stabilization/modification. The majority of these proteins, including a structural maintenance of chromosomes protein (CsSMC), a translin-associated factor X (CsTRAX), a VirE2-interacting protein (CsVIP2), a high mobility group (CsHMG) and two poly(A)-binding proteins (CsPABP1 and 2), interacted with each other, suggesting that they assemble into a multiprotein complex. CsHMG was shown to bind DNA and to interact with the invariable leucine-rich repeat region of PthAs. Surprisingly, both CsHMG and PthA4 interacted with PABP1 and 2 and showed selective binding to poly(U) RNA, a property that is novel among HMGs and TAL effectors. Given that homologs of CsHMG, CsPABP1, CsPABP2, CsSMC and CsTRAX in other organisms assemble into protein complexes to regulate mRNA stability and translation, we suggest a novel role of TAL effectors in mRNA processing and translational control.
Conflict of interest statement
Figures



References
-
- Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010;48:419–436. - PubMed
-
- Kay S, Hahn S, Marois E, Hause G, Bonas U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science. 2007;318:648–651. - PubMed
-
- Römer P, Hahn S, Jordan T, Strauss T, Bonas U, et al. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science. 2007;318:645–648. - PubMed
-
- Kay S, Hahn S, Marois E, Wieduwild R, Bonas U. Detailed analysis of the DNA recognition motifs of the Xanthomonas type III effectors AvrBs3 and AvrBs3Deltarep16. Plant J. 2009;59:859–871. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

