Pivotal role of inosine triphosphate pyrophosphatase in maintaining genome stability and the prevention of apoptosis in human cells
- PMID: 22384212
- PMCID: PMC3288088
- DOI: 10.1371/journal.pone.0032313
Pivotal role of inosine triphosphate pyrophosphatase in maintaining genome stability and the prevention of apoptosis in human cells
Abstract
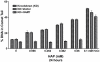
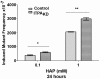
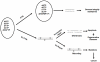
Pure nucleotide precursor pools are a prerequisite for high-fidelity DNA replication and the suppression of mutagenesis and carcinogenesis. ITPases are nucleoside triphosphate pyrophosphatases that clean the precursor pools of the non-canonical triphosphates of inosine and xanthine. The precise role of the human ITPase, encoded by the ITPA gene, is not clearly defined. ITPA is clinically important because a widespread polymorphism, 94C>A, leads to null ITPase activity in erythrocytes and is associated with an adverse reaction to thiopurine drugs. We studied the cellular function of ITPA in HeLa cells using the purine analog 6-N hydroxylaminopurine (HAP), whose triphosphate is also a substrate for ITPA. In this study, we demonstrate that ITPA knockdown sensitizes HeLa cells to HAP-induced DNA breaks and apoptosis. The HAP-induced DNA damage and cytotoxicity observed in ITPA knockdown cells are rescued by an overexpression of the yeast ITPase encoded by the HAM1 gene. We further show that ITPA knockdown results in elevated mutagenesis in response to HAP treatment. Our studies reveal the significance of ITPA in preventing base analog-induced apoptosis, DNA damage and mutagenesis in human cells. This implies that individuals with defective ITPase are predisposed to genome damage by impurities in nucleotide pools, which is drastically augmented by therapy with purine analogs. They are also at an elevated risk for degenerative diseases and cancer.
Conflict of interest statement
Figures
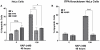
Similar articles
-
ITPA (inosine triphosphate pyrophosphatase): from surveillance of nucleotide pools to human disease and pharmacogenetics.Mutat Res. 2013 Oct-Dec;753(2):131-146. doi: 10.1016/j.mrrev.2013.08.001. Epub 2013 Aug 19. Mutat Res. 2013. PMID: 23969025 Free PMC article. Review.
-
Quantitative in vitro and in vivo characterization of the human P32T mutant ITPase.Biochim Biophys Acta. 2010 Feb;1802(2):269-74. doi: 10.1016/j.bbadis.2009.11.002. Epub 2009 Nov 13. Biochim Biophys Acta. 2010. PMID: 19914375 Free PMC article.
-
RNAi mediated gene silencing of ITPA using a targeted nanocarrier: Apoptosis induction in SKBR3 cancer cells.Clin Exp Pharmacol Physiol. 2017 Aug;44(8):888-894. doi: 10.1111/1440-1681.12776. Clin Exp Pharmacol Physiol. 2017. PMID: 28464292
-
Inosine Triphosphate Pyrophosphatase Dephosphorylates Ribavirin Triphosphate and Reduced Enzymatic Activity Potentiates Mutagenesis in Hepatitis C Virus.J Virol. 2018 Sep 12;92(19):e01087-18. doi: 10.1128/JVI.01087-18. Print 2018 Oct 1. J Virol. 2018. PMID: 30045981 Free PMC article.
-
Inosine Triphosphate Pyrophosphatase (ITPase): Functions, Mutations, Polymorphisms and Its Impact on Cancer Therapies.Cells. 2022 Jan 24;11(3):384. doi: 10.3390/cells11030384. Cells. 2022. PMID: 35159194 Free PMC article. Review.
Cited by
-
The human ITPA polymorphic variant P32T is destabilized by the unpacking of the hydrophobic core.J Struct Biol. 2013 Jun;182(3):197-208. doi: 10.1016/j.jsb.2013.03.007. Epub 2013 Mar 23. J Struct Biol. 2013. PMID: 23528839 Free PMC article.
-
Genetic screens in isogenic mammalian cell lines without single cell cloning.Nat Commun. 2020 Feb 6;11(1):752. doi: 10.1038/s41467-020-14620-6. Nat Commun. 2020. PMID: 32029722 Free PMC article.
-
Viral inosine triphosphatase: A mysterious enzyme with typical activity, but an atypical function.Mol Plant Pathol. 2021 Mar;22(3):382-389. doi: 10.1111/mpp.13021. Epub 2021 Jan 20. Mol Plant Pathol. 2021. PMID: 33471956 Free PMC article. Review.
-
The pivotal role of the mitochondrial amidoxime reducing component 2 in protecting human cells against apoptotic effects of the base analog N6-hydroxylaminopurine.J Biol Chem. 2015 Apr 17;290(16):10126-35. doi: 10.1074/jbc.M115.640052. Epub 2015 Feb 23. J Biol Chem. 2015. PMID: 25713076 Free PMC article.
-
DCTPP1 attenuates the sensitivity of human gastric cancer cells to 5-fluorouracil by up-regulating MDR1 expression epigenetically.Oncotarget. 2016 Oct 18;7(42):68623-68637. doi: 10.18632/oncotarget.11864. Oncotarget. 2016. PMID: 27612427 Free PMC article.
References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
-
- Galperin MY, Moroz OV, Wilson KS, Murzin AG. House cleaning, a part of good housekeeping. Molecular Microbiology. 2006;59:5–19. - PubMed
-
- Mathews CK. DNA precursor metabolism and genomic stability. FASEB J. 2006;20:1300–1314. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

