Differential splicing alters subcellular localization of the alpha but not beta isoform of the MIER1 transcriptional regulator in breast cancer cells
- PMID: 22384264
- PMCID: PMC3286477
- DOI: 10.1371/journal.pone.0032499
Differential splicing alters subcellular localization of the alpha but not beta isoform of the MIER1 transcriptional regulator in breast cancer cells
Abstract
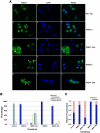
MIER1 was originally identified in a screen for novel fibroblast growth factor activated early response genes. The mier1 gene gives rise to multiple transcripts encoding protein isoforms that differ in their amino (N-) and carboxy (C-) termini. Much of the work to date has focused on the two C-terminal variants, MIER1α and β, both of which have been shown to function as transcriptional repressors. Our previous work revealed a dramatic shift in MIER1α subcellular localization from nuclear in normal breast tissue to cytoplasmic in invasive breast carcinoma, suggesting that loss of nuclear MIER1α may play a role in breast cancer development. In the present study, we investigated whether alternative splicing to include a cassette exon and produce an N-terminal variant of MIER1α affects its subcellular localization in MCF7 breast carcinoma cells. We demonstrate that this cassette exon, exon 3A, encodes a consensus leucine-rich nuclear export signal (NES). Inclusion of this exon in MIER1α to produce the MIER1-3Aα isoform altered its subcellular distribution in MCF7 cells from 81% nuclear to 2% nuclear and this change in localization was abrogated by mutation of critical leucines within the NES. Treatment with leptomycin B (LMB), an inhibitor of the nuclear export receptor CRM1, resulted in a significant increase in the percentage of cells with nuclear MIER1-3Aα, from 4% to 53%, demonstrating that cytoplasmic localization of this isoform was due to CRM1-dependent nuclear export. Inclusion of exon 3A in MIER1β to produce the N-terminal variant MIER1-3Aβ however had little effect on the nuclear targeting of this isoform. Our results demonstrate that alternative splicing to include exon 3A specifically affects the localization pattern of the α isoform.
Conflict of interest statement
Figures



References
-
- Paterno GD, Li Y, Luchman HA, Ryan PJ, Gillespie LL. cDNA cloning of a novel, developmentally regulated immediate early gene activated by fibroblast growth factor and encoding a nuclear protein. J Biol Chem. 1997;272:25591–25595. - PubMed
-
- Paterno GD, Mercer FC, Chayter JJ, Yang X, Robb JD, et al. Molecular cloning of human er1 cDNA and its differential expression in breast tumours and tumour-derived cell lines. Gene. 1998;222:77–82. - PubMed
-
- Paterno GD, Ding Z, Lew YY, Nash GW, Mercer FC, et al. Genomic organization of the human mi-er1 gene and characterization of alternatively spliced isoforms: regulated use of a facultative intron determines subcellular localization. Gene. 2002;295:79–88. - PubMed
-
- Thorne LB, Grant AL, Paterno GD, Gillespie LL. Cloning and characterization of the mouse ortholog of mi-er1. DNA Seq. 2005;16:237–240. - PubMed
-
- Post JN, Gillespie LL, Paterno GD. Nuclear localization signals in the Xenopus FGF embryonic early response 1 protein. FEBS Lett. 2001;502:41–45. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

