The Awesome Power of Yeast Evolutionary Genetics: New Genome Sequences and Strain Resources for the Saccharomyces sensu stricto Genus
- PMID: 22384314
- PMCID: PMC3276118
- DOI: 10.1534/g3.111.000273
The Awesome Power of Yeast Evolutionary Genetics: New Genome Sequences and Strain Resources for the Saccharomyces sensu stricto Genus
Abstract
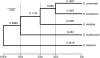
High-quality, well-annotated genome sequences and standardized laboratory strains fuel experimental and evolutionary research. We present improved genome sequences of three species of Saccharomyces sensu stricto yeasts: S. bayanus var. uvarum (CBS 7001), S. kudriavzevii (IFO 1802(T) and ZP 591), and S. mikatae (IFO 1815(T)), and describe their comparison to the genomes of S. cerevisiae and S. paradoxus. The new sequences, derived by assembling millions of short DNA sequence reads together with previously published Sanger shotgun reads, have vastly greater long-range continuity and far fewer gaps than the previously available genome sequences. New gene predictions defined a set of 5261 protein-coding orthologs across the five most commonly studied Saccharomyces yeasts, enabling a re-examination of the tempo and mode of yeast gene evolution and improved inferences of species-specific gains and losses. To facilitate experimental investigations, we generated genetically marked, stable haploid strains for all three of these Saccharomyces species. These nearly complete genome sequences and the collection of genetically marked strains provide a valuable toolset for comparative studies of gene function, metabolism, and evolution, and render Saccharomyces sensu stricto the most experimentally tractable model genus. These resources are freely available and accessible through www.SaccharomycesSensuStricto.org.
Keywords: yeast species; Saccharomyces genome; evolutionary genetics; genome assembly; genomics; sensu stricto.
Figures
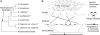

Similar articles
-
Analysis of the constitution of the beer yeast genome by PCR, sequencing and subtelomeric sequence hybridization.Int J Syst Evol Microbiol. 2001 Jul;51(Pt 4):1607-1618. doi: 10.1099/00207713-51-4-1607. Int J Syst Evol Microbiol. 2001. PMID: 11491364
-
Analysis of the genetic variability in the species of the Saccharomyces sensu stricto complex.Yeast. 2003 Oct 30;20(14):1213-26. doi: 10.1002/yea.1034. Yeast. 2003. PMID: 14587104
-
Phylogenetic analysis of the Saccharomyces cerevisiae group based on polymorphisms of rDNA spacer sequences.Int J Syst Bacteriol. 1998 Jan;48 Pt 1:295-303. doi: 10.1099/00207713-48-1-295. Int J Syst Bacteriol. 1998. PMID: 9542100
-
Saccharomyces sensu stricto as a model system for evolution and ecology.Trends Ecol Evol. 2008 Sep;23(9):494-501. doi: 10.1016/j.tree.2008.05.005. Epub 2008 Jul 24. Trends Ecol Evol. 2008. PMID: 18656281 Review.
-
The ecology and evolution of non-domesticated Saccharomyces species.Yeast. 2014 Dec;31(12):449-62. doi: 10.1002/yea.3040. Epub 2014 Oct 23. Yeast. 2014. PMID: 25242436 Free PMC article. Review.
Cited by
-
Negative feedback confers mutational robustness in yeast transcription factor regulation.Proc Natl Acad Sci U S A. 2012 Mar 6;109(10):3874-8. doi: 10.1073/pnas.1116360109. Epub 2012 Feb 21. Proc Natl Acad Sci U S A. 2012. PMID: 22355134 Free PMC article.
-
Most Pleiotropic Effects of Gene Knockouts Are Evolutionarily Transient in Yeasts.Mol Biol Evol. 2024 Sep 4;41(9):msae189. doi: 10.1093/molbev/msae189. Mol Biol Evol. 2024. PMID: 39238468 Free PMC article.
-
Population Size, Sex and Purifying Selection: Comparative Genomics of Two Sister Taxa of the Wild Yeast Saccharomyces paradoxus.Genome Biol Evol. 2020 Sep 1;12(9):1636-1645. doi: 10.1093/gbe/evaa141. Genome Biol Evol. 2020. PMID: 33011797 Free PMC article.
-
Joint inference and alignment of genome structures enables characterization of compartment-independent reorganization across cell types.Epigenetics Chromatin. 2019 Oct 8;12(1):61. doi: 10.1186/s13072-019-0308-3. Epigenetics Chromatin. 2019. PMID: 31594535 Free PMC article.
-
A Unique Saccharomyces cerevisiae × Saccharomyces uvarum Hybrid Isolated From Norwegian Farmhouse Beer: Characterization and Reconstruction.Front Microbiol. 2018 Sep 24;9:2253. doi: 10.3389/fmicb.2018.02253. eCollection 2018. Front Microbiol. 2018. PMID: 30319573 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
