Translational Genomics in Legumes Allowed Placing In Silico 5460 Unigenes on the Pea Functional Map and Identified Candidate Genes in Pisum sativum L
- PMID: 22384322
- PMCID: PMC3276132
- DOI: 10.1534/g3.111.000349
Translational Genomics in Legumes Allowed Placing In Silico 5460 Unigenes on the Pea Functional Map and Identified Candidate Genes in Pisum sativum L
Abstract
To identify genes involved in phenotypic traits, translational genomics from highly characterized model plants to poorly characterized crop plants provides a valuable source of markers to saturate a zone of interest as well as functionally characterized candidate genes. In this paper, an integrated view of the pea genetic map was developed. A series of gene markers were mapped and their best reciprocal homologs were identified on M. truncatula, L. japonicus, soybean, and poplar pseudomolecules. Based on the syntenic relationships uncovered between pea and M. truncatula, 5460 pea Unigenes were tentatively placed on the consensus map. A new bioinformatics tool, http://www.thelegumeportal.net/pea_mtr_translational_toolkit, was developed that allows, for any gene sequence, to search its putative position on the pea consensus map and hence to search for candidate genes among neighboring Unigenes. As an example, a promising candidate gene for the hypernodulation mutation nod3 in pea was proposed based on the map position of the likely homolog of Pub1, a M. truncatula gene involved in nodulation regulation. A broader view of pea genome evolution was obtained by revealing syntenic relationships between pea and sequenced genomes. Blocks of synteny were identified which gave new insights into the evolution of chromosome structure in Papillionoids and Eudicots. The power of the translational genomics approach was underlined.
Keywords: Pisum sativum; functional consensus map; model legume species; synteny; translational genomics.
Figures



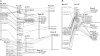

References
-
- Abrouk M., Murat F., Pont C., Messing J., Jackson S., et al. , 2010. Palaeogenomics of plants: synteny-based modelling of extinct ancestors. Trends Plant Sci. 15: 479–487 - PubMed
-
- Aubert G., Morin J., Jacquin J., Loridon K., Quillet M. C., et al. , 2006. Functional mapping in pea, as an aid to the candidate gene selection and for investigating synteny with the model legume Medicago truncatula. Theor. Appl. Genet. 112: 1024–1041 - PubMed
-
- Blixt S., 1972. Mutation genetics in Pisum. Agri. Hort. Genet. 30: 1–293
LinkOut - more resources
Full Text Sources
Research Materials
