High-Resolution Genotyping of Wild Barley Introgression Lines and Fine-Mapping of the Threshability Locus thresh-1 Using the Illumina GoldenGate Assay
- PMID: 22384330
- PMCID: PMC3276139
- DOI: 10.1534/g3.111.000182
High-Resolution Genotyping of Wild Barley Introgression Lines and Fine-Mapping of the Threshability Locus thresh-1 Using the Illumina GoldenGate Assay
Abstract
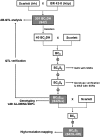
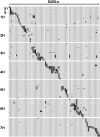
Genetically well-characterized mapping populations are a key tool for rapid and precise localization of quantitative trait loci (QTL) and subsequent identification of the underlying genes. In this study, a set of 73 introgression lines (S42ILs) originating from a cross between the spring barley cultivar Scarlett (Hordeum vulgare ssp. vulgare) and the wild barley accession ISR42-8 (H. v. ssp. spontaneum) was subjected to high-resolution genotyping with an Illumina 1536-SNP array. The array enabled a precise localization of the wild barley introgressions in the elite barley background. Based on 636 informative SNPs, the S42IL set represents 87.3% of the wild barley genome, where each line contains on average 3.3% of the donor genome. Furthermore, segregating high-resolution mapping populations (S42IL-HRs) were developed for 70 S42ILs in order to facilitate QTL fine-mapping and cloning. As a case study, we used the developed genetic resources to rapidly identify and fine-map the novel locus thresh-1 on chromosome 1H that controls grain threshability. Here, the recessive wild barley allele confers a difficult to thresh phenotype, suggesting that thresh-1 played an important role during barley domestication. Using a S42IL-HR population, thresh-1 was fine-mapped within a 4.3cM interval that was predicted to contain candidate genes involved in regulation of plant cell wall composition. The set of wild barley introgression lines and derived high-resolution populations are ideal tools to speed up the process of mapping and further dissecting QTL, which ultimately clears the way for isolating the genes behind QTL effects.
Keywords: Illumina GoldenGate assay; high-resolution mapping populations; threshability locus thresh-1; wild barley introgression lines.
Figures


Similar articles
-
Identification and verification of QTLs for agronomic traits using wild barley introgression lines.Theor Appl Genet. 2009 Feb;118(3):483-97. doi: 10.1007/s00122-008-0915-z. Epub 2008 Nov 1. Theor Appl Genet. 2009. PMID: 18979081
-
Detection and verification of malting quality QTLs using wild barley introgression lines.Theor Appl Genet. 2009 May;118(8):1411-27. doi: 10.1007/s00122-009-0991-8. Epub 2009 Mar 3. Theor Appl Genet. 2009. PMID: 19255740 Free PMC article.
-
Wild barley introgression lines revealed novel QTL alleles for root and related shoot traits in the cultivated barley (Hordeum vulgare L.).BMC Genet. 2014 Oct 7;15:107. doi: 10.1186/s12863-014-0107-6. BMC Genet. 2014. PMID: 25286820 Free PMC article.
-
Association of barley photoperiod and vernalization genes with QTLs for flowering time and agronomic traits in a BC2DH population and a set of wild barley introgression lines.Theor Appl Genet. 2010 May;120(8):1559-74. doi: 10.1007/s00122-010-1276-y. Epub 2010 Feb 13. Theor Appl Genet. 2010. PMID: 20155245 Free PMC article.
-
Selecting a set of wild barley introgression lines and verification of QTL effects for resistance to powdery mildew and leaf rust.Theor Appl Genet. 2008 Nov;117(7):1093-106. doi: 10.1007/s00122-008-0847-7. Epub 2008 Jul 29. Theor Appl Genet. 2008. PMID: 18663425
Cited by
-
Global Transcriptome Profiling of Developing Leaf and Shoot Apices Reveals Distinct Genetic and Environmental Control of Floral Transition and Inflorescence Development in Barley.Plant Cell. 2015 Sep;27(9):2318-34. doi: 10.1105/tpc.15.00203. Epub 2015 Aug 25. Plant Cell. 2015. PMID: 26307377 Free PMC article.
-
Modelling the genetic architecture of flowering time control in barley through nested association mapping.BMC Genomics. 2015 Apr 12;16(1):290. doi: 10.1186/s12864-015-1459-7. BMC Genomics. 2015. PMID: 25887319 Free PMC article.
-
Emerging paradigms in genomics-based crop improvement.ScientificWorldJournal. 2013 Nov 17;2013:585467. doi: 10.1155/2013/585467. ScientificWorldJournal. 2013. PMID: 24348171 Free PMC article. Review.
-
Photoperiod-H1 (Ppd-H1) Controls Leaf Size.Plant Physiol. 2016 Sep;172(1):405-15. doi: 10.1104/pp.16.00977. Epub 2016 Jul 25. Plant Physiol. 2016. PMID: 27457126 Free PMC article.
-
Genetic mapping of a barley leaf rust resistance gene Rph26 introgressed from Hordeum bulbosum.Theor Appl Genet. 2018 Dec;131(12):2567-2580. doi: 10.1007/s00122-018-3173-8. Epub 2018 Sep 3. Theor Appl Genet. 2018. PMID: 30178277
References
-
- Azhacuvel P., Vidya-Saraswathi D., Komatsuda T., 2006. High-resolution linkage mapping for the non-brittle rachis locus btr1 in cultivated x wild barley (Hordeum vulgare). Plant Sci. 170: 1087–1094
-
- Chin J. H., Lu X., Haefele S. M., Gamuyao R., Ismail A., et al. , 2010. Development and application of gene-based markers for the major rice QTL Phosphorus uptake 1. Theor. Appl. Genet. 120: 1073–1086 - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
