Genotype imputation with thousands of genomes
- PMID: 22384356
- PMCID: PMC3276165
- DOI: 10.1534/g3.111.001198
Genotype imputation with thousands of genomes
Abstract
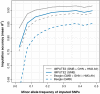
Genotype imputation is a statistical technique that is often used to increase the power and resolution of genetic association studies. Imputation methods work by using haplotype patterns in a reference panel to predict unobserved genotypes in a study dataset, and a number of approaches have been proposed for choosing subsets of reference haplotypes that will maximize accuracy in a given study population. These panel selection strategies become harder to apply and interpret as sequencing efforts like the 1000 Genomes Project produce larger and more diverse reference sets, which led us to develop an alternative framework. Our approach is built around a new approximation that uses local sequence similarity to choose a custom reference panel for each study haplotype in each region of the genome. This approximation makes it computationally efficient to use all available reference haplotypes, which allows us to bypass the panel selection step and to improve accuracy at low-frequency variants by capturing unexpected allele sharing among populations. Using data from HapMap 3, we show that our framework produces accurate results in a wide range of human populations. We also use data from the Malaria Genetic Epidemiology Network (MalariaGEN) to provide recommendations for imputation-based studies in Africa. We demonstrate that our approximation improves efficiency in large, sequence-based reference panels, and we discuss general computational strategies for modern reference datasets. Genome-wide association studies will soon be able to harness the power of thousands of reference genomes, and our work provides a practical way for investigators to use this rich information. New methodology from this study is implemented in the IMPUTE2 software package.
Keywords: GWAS; haplotype; human; linkage disequilibrium; reference panel.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
