NMR determination of protein partitioning into membrane domains with different curvatures and application to the influenza M2 peptide
- PMID: 22385849
- PMCID: PMC3283775
- DOI: 10.1016/j.bpj.2012.01.010
NMR determination of protein partitioning into membrane domains with different curvatures and application to the influenza M2 peptide
Abstract
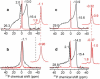
The M2 protein of the influenza A virus acts both as a drug-sensitive proton channel and mediates virus budding through membrane scission. The segment responsible for causing membrane curvature is an amphipathic helix in the cytoplasmic domain of the protein. Here, we use (31)P and (13)C solid-state NMR to examine M2-induced membrane curvature. M2(22-46), which includes only the transmembrane (TM) helix, and M2(21-61), which contains an additional amphipathic helix, are studied. (31)P chemical shift lineshapes indicate that M2(21-61) causes a high-curvature isotropic phase to both cholesterol-rich virus-mimetic membranes and 1,2-dimyristoyl-sn-glycero-3-phosphocholine bilayers, whereas M2(22-46) has minimal effect. The lamellar and isotropic domains have distinct (31)P isotropic chemical shifts, indicating perturbation of the lipid headgroup conformation by the amphipathic helix. (31)P- and (13)C-detected (1)H T(2) relaxation and two-dimensional peptide-lipid correlation spectra show that M2(21-61) preferentially binds to the high-curvature domain. (31)P linewidths indicate that the isotropic vesicles induced by M2(21-61) are 10-35 nm in diameter, and the virus-mimetic vesicles are smaller than the 1,2-dimyristoyl-sn-glycero-3-phosphocholine vesicles. A strong correlation is found between high membrane curvature and weak drug-binding ability of the TM helix. Thus, the M2 amphipathic helix causes membrane curvature, which in turn perturbs the TM helix conformation, abolishing drug binding. These NMR experiments are applicable to other curvature-inducing membrane proteins such as fusion proteins and antimicrobial peptides.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Investigation of the curvature induction and membrane localization of the influenza virus M2 protein using static and off-magic-angle spinning solid-state nuclear magnetic resonance of oriented bicelles.Biochemistry. 2015 Apr 7;54(13):2214-26. doi: 10.1021/acs.biochem.5b00127. Epub 2015 Mar 26. Biochemistry. 2015. PMID: 25774685 Free PMC article.
-
Isotropic bicelles stabilize the juxtamembrane region of the influenza M2 protein for solution NMR studies.Biochemistry. 2013 Nov 26;52(47):8420-9. doi: 10.1021/bi401035m. Epub 2013 Nov 14. Biochemistry. 2013. PMID: 24168642
-
Cholesterol-binding site of the influenza M2 protein in lipid bilayers from solid-state NMR.Proc Natl Acad Sci U S A. 2017 Dec 5;114(49):12946-12951. doi: 10.1073/pnas.1715127114. Epub 2017 Nov 20. Proc Natl Acad Sci U S A. 2017. PMID: 29158386 Free PMC article.
-
Structural basis for proton conduction and inhibition by the influenza M2 protein.Protein Sci. 2012 Nov;21(11):1620-33. doi: 10.1002/pro.2158. Epub 2012 Oct 9. Protein Sci. 2012. PMID: 23001990 Free PMC article. Review.
-
Cholesterol-binding viral proteins in virus entry and morphogenesis.Subcell Biochem. 2010;51:77-108. doi: 10.1007/978-90-481-8622-8_3. Subcell Biochem. 2010. PMID: 20213541 Free PMC article. Review.
Cited by
-
Magic angle spinning NMR of viruses.Prog Nucl Magn Reson Spectrosc. 2015 Apr;86-87:21-40. doi: 10.1016/j.pnmrs.2015.02.003. Epub 2015 Feb 16. Prog Nucl Magn Reson Spectrosc. 2015. PMID: 25919197 Free PMC article. Review.
-
Cholesterol and M2 Rendezvous in Budding and Scission of Influenza A Virus.Subcell Biochem. 2023;106:441-459. doi: 10.1007/978-3-031-40086-5_16. Subcell Biochem. 2023. PMID: 38159237
-
Spherical nanoparticle supported lipid bilayers for the structural study of membrane geometry-sensitive molecules.J Am Chem Soc. 2015 Nov 11;137(44):14031-14034. doi: 10.1021/jacs.5b08303. Epub 2015 Oct 28. J Am Chem Soc. 2015. PMID: 26488086 Free PMC article.
-
Structural Basis for Asymmetric Conductance of the Influenza M2 Proton Channel Investigated by Solid-State NMR Spectroscopy.J Mol Biol. 2017 Jul 7;429(14):2192-2210. doi: 10.1016/j.jmb.2017.05.015. Epub 2017 May 20. J Mol Biol. 2017. PMID: 28535993 Free PMC article.
-
Entropic forces drive clustering and spatial localization of influenza A M2 during viral budding.Proc Natl Acad Sci U S A. 2018 Sep 11;115(37):E8595-E8603. doi: 10.1073/pnas.1805443115. Epub 2018 Aug 27. Proc Natl Acad Sci U S A. 2018. PMID: 30150411 Free PMC article.
References
-
- Han X., Bushweller J.H., Tamm L.K. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 2001;8:715–720. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

