Minimalist model for force-dependent DNA replication
- PMID: 22385852
- PMCID: PMC3283810
- DOI: 10.1016/j.bpj.2012.01.020
Minimalist model for force-dependent DNA replication
Abstract
In experiments using optical or magnetic tweezers, investigators have monitored the rate at which polymerase enzymes catalyze DNA replication when the template strand is subjected to a stretching force. For T7, Klenow, and Sequenase polymerases, the replication rate increases modestly at low tension and then decreases markedly at higher tension. Molecular-dynamics (MD) simulations using x-ray structure data for the open and closed complexes of the Taq enzyme with DNA revealed that the dependence of replication rate on tension could be accounted for in terms of the induced enthalpy changes for the two DNA segments adjacent to the site of the added nucleotide. Here, we present a simple, minimalist two-segment local model (M2L) derived from some striking features seen in the MD simulations. The model predicts the tension dependence of the replication rate using only structural data and a critical tension, f(∗), without recourse to MD simulations. At f(∗), the outermost DNA segment undergoes a large angular reorientation in the open conformation of the enzyme. We give a generic plot for the M2L model, apply it to family A and B polymerases and HIV reverse transcriptase, and discuss factors that may govern the f(∗) flip parameter.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

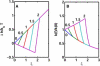
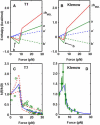
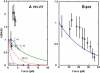

Similar articles
-
Tuning DNA "strings": modulating the rate of DNA replication with mechanical tension.Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8485-9. doi: 10.1073/pnas.151261198. Epub 2001 Jul 10. Proc Natl Acad Sci U S A. 2001. PMID: 11447284 Free PMC article.
-
Unifying themes in DNA replication: reconciling single molecule kinetic studies with structural data on DNA polymerases.J Biomol Struct Dyn. 2002 Feb;19(4):571-84. doi: 10.1080/07391102.2002.10506764. J Biomol Struct Dyn. 2002. PMID: 11843619
-
Insights into DNA polymerization mechanisms from structure and function analysis of HIV-1 reverse transcriptase.Biochemistry. 1995 Apr 25;34(16):5351-63. doi: 10.1021/bi00016a006. Biochemistry. 1995. PMID: 7537090
-
Protein-nucleic acid interactions and DNA conformation in a complex of human immunodeficiency virus type 1 reverse transcriptase with a double-stranded DNA template-primer.Biopolymers. 1997;44(2):125-38. doi: 10.1002/(SICI)1097-0282(1997)44:2<125::AID-BIP2>3.0.CO;2-X. Biopolymers. 1997. PMID: 9354757 Review.
-
Interaction of retroviral reverse transcriptase with template-primer duplexes during replication.Prog Nucleic Acid Res Mol Biol. 1998;58:339-93. doi: 10.1016/s0079-6603(08)60041-0. Prog Nucleic Acid Res Mol Biol. 1998. PMID: 9308371 Review.
Cited by
-
DNA replication: In vitro single-molecule manipulation data analysis and models.Comput Struct Biotechnol J. 2021 Jun 24;19:3765-3778. doi: 10.1016/j.csbj.2021.06.032. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34285777 Free PMC article. Review.
-
A DNA-centered explanation of the DNA polymerase translocation mechanism.Sci Rep. 2017 Aug 8;7(1):7566. doi: 10.1038/s41598-017-08038-2. Sci Rep. 2017. PMID: 28790383 Free PMC article.
-
Replicative DNA polymerases promote active displacement of SSB proteins during lagging strand synthesis.Nucleic Acids Res. 2019 Jun 20;47(11):5723-5734. doi: 10.1093/nar/gkz249. Nucleic Acids Res. 2019. PMID: 30968132 Free PMC article.
References
-
- Wuite G.J., Smith S.B., Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404:103–106. - PubMed
-
- Smith D.E., Tans S.J., Bustamante C. The bacteriophage straight ϕ29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. - PubMed
-
- Joyce C.M., Steitz T.A. Function and structure relationships in DNA Polymerases. Annu. Rev. Biochem. 1994;63:777–822. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

