RNA and its ionic cloud: solution scattering experiments and atomically detailed simulations
- PMID: 22385853
- PMCID: PMC3283807
- DOI: 10.1016/j.bpj.2012.01.013
RNA and its ionic cloud: solution scattering experiments and atomically detailed simulations
Abstract
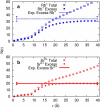
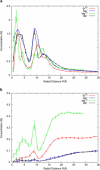
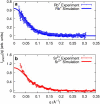
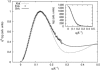
RNA molecules play critical roles in many cellular processes. Traditionally viewed as genetic messengers, RNA molecules were recently discovered to have diverse functions related to gene regulation and expression. RNA also has great potential as a therapeutic and a tool for further investigation of gene regulation. Metal ions are an integral part of RNA structure and should be considered in any experimental or theoretical study of RNA. Here, we report a multidisciplinary approach that combines anomalous small-angle x-ray scattering and molecular-dynamics (MD) simulations with explicit solvent and ions around RNA. From experiment and simulation results, we find excellent agreement in the number and distribution of excess monovalent and divalent ions around a short RNA duplex. Although similar agreement can be obtained from a continuum description of the solvent and mobile ions (by solving the Poisson-Boltzmann equation and accounting for finite ion size), the use of MD is easily extended to flexible RNA systems with thermal fluctuations. Therefore, we also model a short RNA pseudoknot and find good agreement between the MD results and the experimentally derived solution structures. Surprisingly, both deviate from crystal structure predictions. These favorable comparisons of experiment and simulations encourage work on RNA in all-atom dynamic models.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Computational exploration of mobile ion distributions around RNA duplex.J Phys Chem B. 2010 Jun 24;114(24):8207-20. doi: 10.1021/jp911992t. J Phys Chem B. 2010. PMID: 20518549 Free PMC article.
-
Reweighting of molecular simulations with explicit-solvent SAXS restraints elucidates ion-dependent RNA ensembles.Nucleic Acids Res. 2021 Aug 20;49(14):e84. doi: 10.1093/nar/gkab459. Nucleic Acids Res. 2021. PMID: 34107023 Free PMC article.
-
Mono- and trivalent ions around DNA: a small-angle scattering study of competition and interactions.Biophys J. 2008 Jul;95(1):287-95. doi: 10.1529/biophysj.107.123174. Epub 2008 Mar 13. Biophys J. 2008. PMID: 18339743 Free PMC article.
-
Hybrid Methods for Modeling Protein Structures Using Molecular Dynamics Simulations and Small-Angle X-Ray Scattering Data.Adv Exp Med Biol. 2018;1105:237-258. doi: 10.1007/978-981-13-2200-6_15. Adv Exp Med Biol. 2018. PMID: 30617833 Review.
-
SAXS studies of ion-nucleic acid interactions.Annu Rev Biophys. 2011;40:225-42. doi: 10.1146/annurev-biophys-042910-155349. Annu Rev Biophys. 2011. PMID: 21332357 Review.
Cited by
-
Computational Study of the pH-Dependent Ionic Environment around β-Lactoglobulin.J Phys Chem B. 2022 Nov 17;126(45):9123-9136. doi: 10.1021/acs.jpcb.2c03797. Epub 2022 Nov 2. J Phys Chem B. 2022. PMID: 36321840 Free PMC article.
-
The ionic atmosphere around A-RNA: Poisson-Boltzmann and molecular dynamics simulations.Biophys J. 2012 Feb 22;102(4):829-38. doi: 10.1016/j.bpj.2011.12.055. Epub 2012 Feb 21. Biophys J. 2012. PMID: 22385854 Free PMC article.
-
DNA A-tracts are not curved in solutions containing high concentrations of monovalent cations.Biochemistry. 2013 Jun 18;52(24):4138-48. doi: 10.1021/bi400118m. Epub 2013 Jun 6. Biochemistry. 2013. PMID: 23675817 Free PMC article.
-
Aggregates Sealed by Ions.Methods Mol Biol. 2022;2340:309-341. doi: 10.1007/978-1-0716-1546-1_14. Methods Mol Biol. 2022. PMID: 35167080
-
Multiscale Multiphysics and Multidomain Models I: Basic Theory.J Theor Comput Chem. 2013 Dec;12(8):10.1142/S021963361341006X. doi: 10.1142/S021963361341006X. J Theor Comput Chem. 2013. PMID: 25382892 Free PMC article.
References
-
- Eddy S.R. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001;2:919–929. - PubMed
-
- Mattick J.S., Makunin I.V. Non-coding RNA. Hum. Mol. Genet. 2006;15(Spec No 1):R17–R29. - PubMed
-
- Chin K., Sharp K.A., Pyle A.M. Calculating the electrostatic properties of RNA provides new insights into molecular interactions and function. Nat. Struct. Biol. 1999;6:1055–1061. - PubMed
-
- Manning G.S. Limiting laws and counterion condensation in poly-electrolyte solutions 8. Mixtures of counterions, specific selectivity, and valence selectivity. J. Phys. Chem. 1984;88:6654–6661.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

