Using DNA as a fiducial marker to study SMC complex interactions with the atomic force microscope
- PMID: 22385855
- PMCID: PMC3283809
- DOI: 10.1016/j.bpj.2012.01.022
Using DNA as a fiducial marker to study SMC complex interactions with the atomic force microscope
Abstract
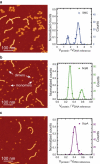
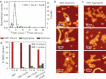
Atomic force microscopy can potentially provide information on protein volumes, shapes, and interactions but is susceptible to variable tip-induced artifacts. In this study, we present an atomic force microscopy approach that can measure volumes of nonglobular polypeptides such as structural maintenance of chromosomes (SMC) proteins, and use it to study the interactions that occur within and between SMC complexes. Together with the protein of interest, we coadsorb a DNA molecule and use it as a fiducial marker to account for tip-induced artifacts that affect both protein and DNA, allowing normalization of protein volumes from images taken on different days and with different tips. This approach significantly reduced the error associated with volume analysis, and allowed determination of the oligomeric states and architecture of the Bacillus subtilis SMC complex, formed by the SMC protein, and by the smaller ScpA and ScpB subunits. This work reveals that SMC and ScpB are dimers and that ScpA is a stable monomer. Moreover, whereas ScpA binds directly to SMC, ScpB only binds to SMC in the presence of ScpA. Notably, the presence of both ScpA and ScpB favored the formation of higher-order structures of SMC complexes, suggesting a role for these subunits in the organization of SMC oligomers.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Binnig G., Quate C.F., Gerber C. Atomic force microscope. Phys. Rev. Lett. 1986;56:930–933. - PubMed
-
- Bustamante C., Keller D. Scanning force microscopy in biology. Phys. Today. 1995;48:32–38.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

