PhiSiGns: an online tool to identify signature genes in phages and design PCR primers for examining phage diversity
- PMID: 22385976
- PMCID: PMC3314551
- DOI: 10.1186/1471-2105-13-37
PhiSiGns: an online tool to identify signature genes in phages and design PCR primers for examining phage diversity
Abstract
Background: Phages (viruses that infect bacteria) have gained significant attention because of their abundance, diversity and important ecological roles. However, the lack of a universal gene shared by all phages presents a challenge for phage identification and characterization, especially in environmental samples where it is difficult to culture phage-host systems. Homologous conserved genes (or "signature genes") present in groups of closely-related phages can be used to explore phage diversity and define evolutionary relationships amongst these phages. Bioinformatic approaches are needed to identify candidate signature genes and design PCR primers to amplify those genes from environmental samples; however, there is currently no existing computational tool that biologists can use for this purpose.
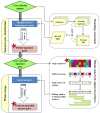
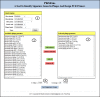
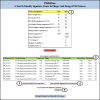
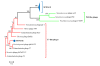
Results: Here we present PhiSiGns, a web-based and standalone application that performs a pairwise comparison of each gene present in user-selected phage genomes, identifies signature genes, generates alignments of these genes, and designs potential PCR primer pairs. PhiSiGns is available at (http://www.phantome.org/phisigns/; http://phisigns.sourceforge.net/) with a link to the source code. Here we describe the specifications of PhiSiGns and demonstrate its application with a case study.
Conclusions: PhiSiGns provides phage biologists with a user-friendly tool to identify signature genes and design PCR primers to amplify related genes from uncultured phages in environmental samples. This bioinformatics tool will facilitate the development of novel signature genes for use as molecular markers in studies of phage diversity, phylogeny, and evolution.
Figures
References
-
- Breitbart M, Thompson L, Suttle C, Sullivan M. Exploring the vast diversity of marine viruses. Oceanography. 2007;20:135–139.
-
- Weinbauer MG, Rassoulzadegan F. Are viruses driving microbial diversification and diversity? Environ Microbiol. 2004;6:1–11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

