Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers
- PMID: 22387464
- PMCID: PMC3384321
- DOI: 10.1093/nar/gks199
Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers
Abstract
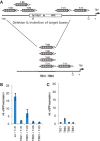
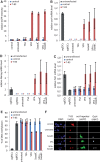
Specific control of gene activity is a valuable tool to study and engineer cellular functions. Recent studies uncovered the potential of transcription activator-like effector (TALE) proteins that can be tailored to activate user-defined target genes. It remains however unclear whether and how epigenetic modifications interfere with TALE-mediated transcriptional activation. We studied the activity of five designer TALEs (dTALEs) targeting the oct4 pluripotency gene. In vitro assays showed that the five dTALEs that target distinct sites in the oct4 promoter had the expected DNA specificity and comparable affinities to their corresponding DNA targets. In contrast to their similar in vitro properties, transcriptional activation of oct4 by these distinct dTALEs varied up to 25-fold. While dTALEs efficiently upregulated transcription of the active oct4 promoter in embryonic stem cells (ESCs) they failed to activate the silenced oct4 promoter in ESC-derived neural stem cells (NSCs), indicating that as for endogenous transcription factors also dTALE activity is limited by repressive epigenetic mechanisms. We therefore targeted the activity of epigenetic modulators and found that chemical inhibition of histone deacetylases by valproic acid or DNA methyltransferases by 5-aza-2'-deoxycytidine facilitated dTALE-mediated activation of the epigenetically silenced oct4 promoter in NSCs. Notably, demethylation of the oct4 promoter occurred only if chemical inhibitors and dTALEs were applied together but not upon treatment with inhibitors or dTALEs only. These results show that dTALEs in combination with chemical manipulation of epigenetic modifiers facilitate targeted transcriptional activation of epigenetically silenced target genes.
Figures


 .
.
References
-
- Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q. Rev. Biophys. 2010;43:1–21. - PubMed
-
- Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Mol. Ther. 2008;16:1200–1207. - PubMed
-
- De Francesco L. Move over ZFNs. Nat. Biotechnol. 2011;29:681–684. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

